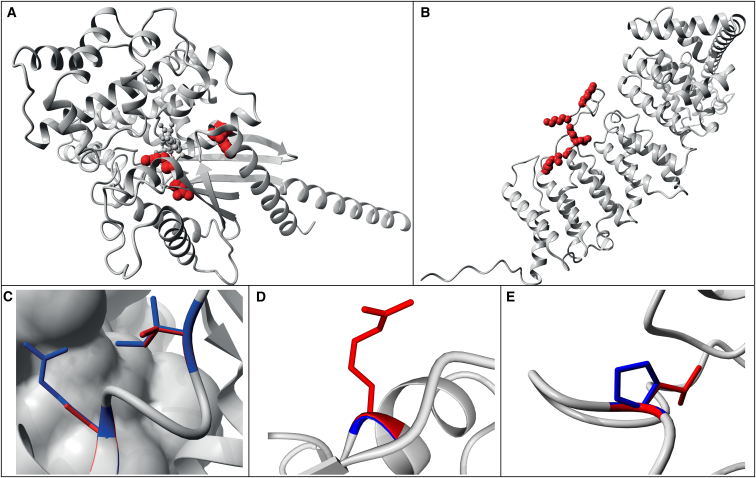
Figure 3.
Examples of Modeling of Missense Mutations on 3D Protein Structures
Wild-type residues are marked in blue; de novo mutations are indicated as red globes or lines (Tables S17).
(A) 3D structure of GNA1, acting through HI, showing that the modeled missense mutations are buried and likely to disrupt protein folding.
(B) Structure of PPP2R5D, acting through NHI, where the modeled missense mutations affect mostly surface residues and are expected to have no or only local structural effects.
(C) Zoom-in of known missense variants p.Arg496Cys and p.Ile500Val in SMAD4 known to act through a gain-of-function mechanism. These variants are located on the surface of the monomer and in contact with another SMAD4 monomer.38
(D) Zoom-in of the missense variant p.Gly343Arg in ACTL6B which is located at the surface. The side-chain points toward the solvent, therefore the larger Arginine will fit.
(E) Zoom-in of the missense variant p.Pro65Leu in PCGF2 close to the interaction site with other molecules.