Figure 3. Reducing Protein Synthesis Increases the Level of UGA Readthrough.
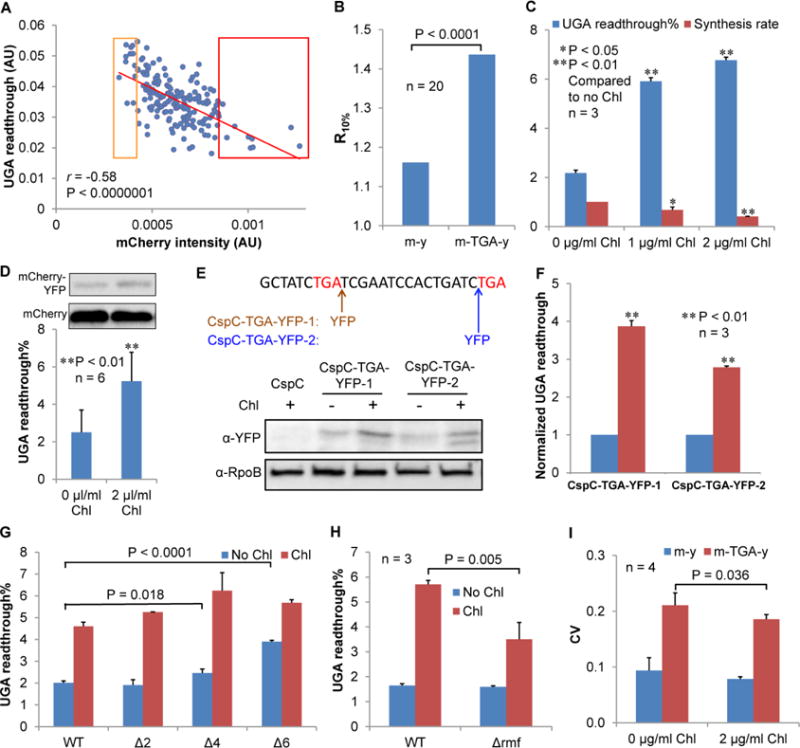
(A) In single MG1655 cells, higher UGA readthrough correlates (Spearman’s Rank correlation) with lower mCherry protein level. The mCherry intensity is the mCherry signal normalized by cell volume with arbitrary units. Relative UGA readthrough is calculated from the YFP/mCherry ratio. Experimental conditions were the same as in Figure 2. (B) The Y-axis (R10%) is the ratio of the average UGA readthrough in the bottom 10% of cells divided by the average UGA readthrough in the top 10% of cells ranked by mCherry intensity from panel A. (C) MG1655 cells were grown in LB with and without Chl for 24 h, and UGA readthrough and protein synthesis rates were determined by fluorescence spectrometry. Treating MG1655 with low doses of Chl decreases protein synthesis rate and enhances UGA readthrough. (D) Western blotting confirms that Chl increases UGA readthrough of the m-TGA-y reporter. (E, F) UGA readthrough of chromosomally-encoded CspC increases in the presence of chloramphenicol. The nucleotide sequence of the 3′-end of the chromosomal constitutively expressed cspC gene is shown. Red letters indicate the first and second stop codons. An in-frame YFP tag is inserted immediately after the first TGA codon of the native cspC gene (CspC-TGA-YFP-1) or before the second stop codon (CspC-TGA-YFP-2) in MG1655. Western blotting result shows that readthrough of the cspC UGA codon is significantly increased by addition of Chl. (G) Reducing ribosome copy number increases UGA readthrough. WT MG1655 and its ribosomal operon deletion mutants (Δ2, ΔrrnEG; Δ4, ΔrrnGBAD; Δ6, ΔrrnGADBHC) were grown in LB with and without Chl for 24 h, and UGA readthrough levels were determined with fluorescence spectrometry. (H) Deleting rmf partially suppresses the effect of Chl to increase UGA readthrough. (I) Chl treatment decreases the CV of UGA readthrough. Data are represented as mean ± standard deviation. AU, arbitrary units.
