Figure 7. Model for UGA Readthrough Heterogeneity.
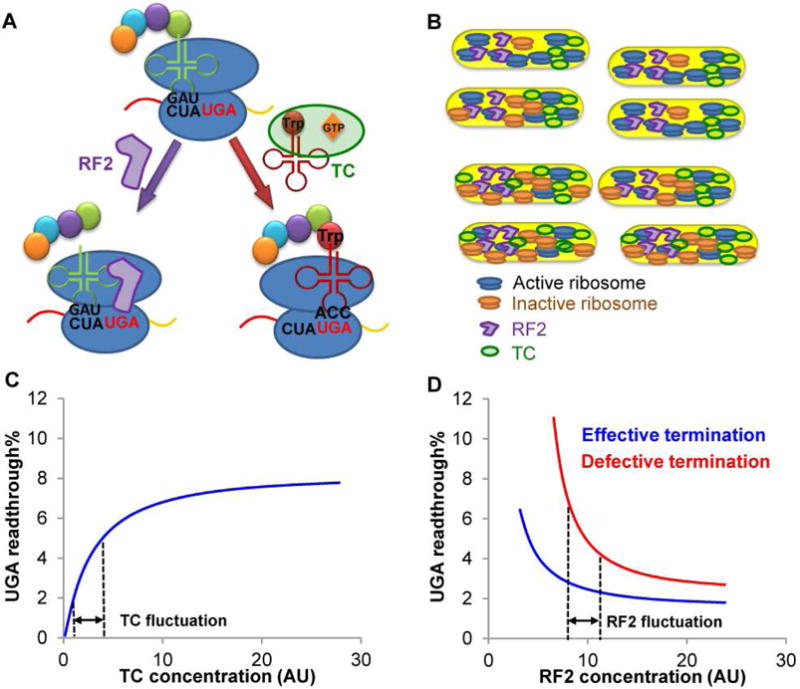
(A) The near-cognate Trp-tRNATrp forms a ternary complex (TC) with EF-Tu and GTP to compete with RF2 for the UGA stop codon. Recognition of UGA by RF2 leads to release of the growing peptide from the ribosome, whereas Trp-tRNATrp suppresses UGA by adding Trp to the growing peptide. The UGA readthrough level in a single cell is determined by the effective concentrations of TC and RF2. (B) The concentration of active ribosomes may vary among single cells due to fluctuations of global resources and ribosome maturation/inactivation. A low concentration of active ribosomes decreases global protein synthesis, which increases the effective concentration of TC due to its reduced usage during translation. (C) UGA readthrough is sensitive to TC fluctuations. An increase in effective TC leads to enhanced UGA readthrough. (D) WT cells with effective translation termination exhibit nearly saturated RF2 activity, and UGA readthrough is insensitive to RF2 fluctuations. Defects in RF2 activity, e.g., due to RF3 deletion, enhance the sensitivity of UGA readthrough to RF2 fluctuations and thus increase UGA readthrough heterogeneity. Dashed lines indicate arbitrary ranges of concentrations. AU, arbitrary units.
