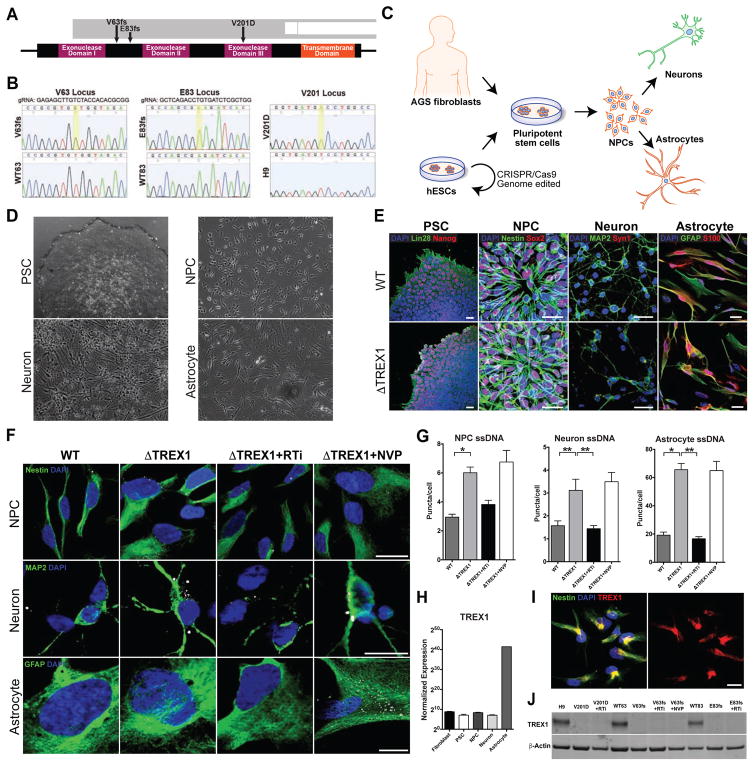
Figure 1. TREX-1-deficient neural cells exhibit higher levels of ssDNA in the cytosol (see also Figures S1–S3).
A, Schematic representation of the TREX1 gene showing the mutations in the derived pluripotent lines. B, DNA sequence chromatogram displaying the nucleotide changes in the TREX1 sequence in the mutant lines. A golden box denotes a nucleotide mutation. The amino acid sequence is provided in the white ribbon beneath the nucleic acid sequence. C, Schematic representation of the generation of cell lines and differentiation into neural cells. D, Phase-contrast images showing differentiation in the neural lineage. Scale bar, 200μm. E, Representative fluorescence images showing differentiation in the neural lineage. Scale bar, 20μm. F, Representative images of ssDNA immunofluorescence in NPCs, neurons and astrocytes. Scale bar, 20μm. G, Quantification of ssDNA puncta in the cytosol of NPCs, neurons and astrocytes. All ssDNA images were blindly acquired and ssDNA puncta were blindly quantified. The numbers of puncta per cell for each line were averaged and graphed according to genotype (n = 3 cell lines). Each mutant line was chronically treated with nucleoside analogue reverse-transcriptase inhibitors 3TC and d4T (RTi), then subjected to imaging, and the results were graphed (n = 3 cell lines). The V63fs line was also chronically treated with the non-nucleoside analogue reverse-transcriptase inhibitor NVP (n = 1 cell line). The presented values are the means ± SD. Student’s t-tests with Welch’s correction were performed to compare genotypes. *P<0.05; **P<0.01. H, Normalized TREX1 expression across differentiation using RNA sequencing data from control cell lines (n = 3 cell lines). The presented values are the means ± SEM. I, Fluorescence images showing subcellular localization of TREX1 in control NPCs. Scale bar, 20μm. J, Western blot on astrocyte protein lysates measuring levels of TREX1.