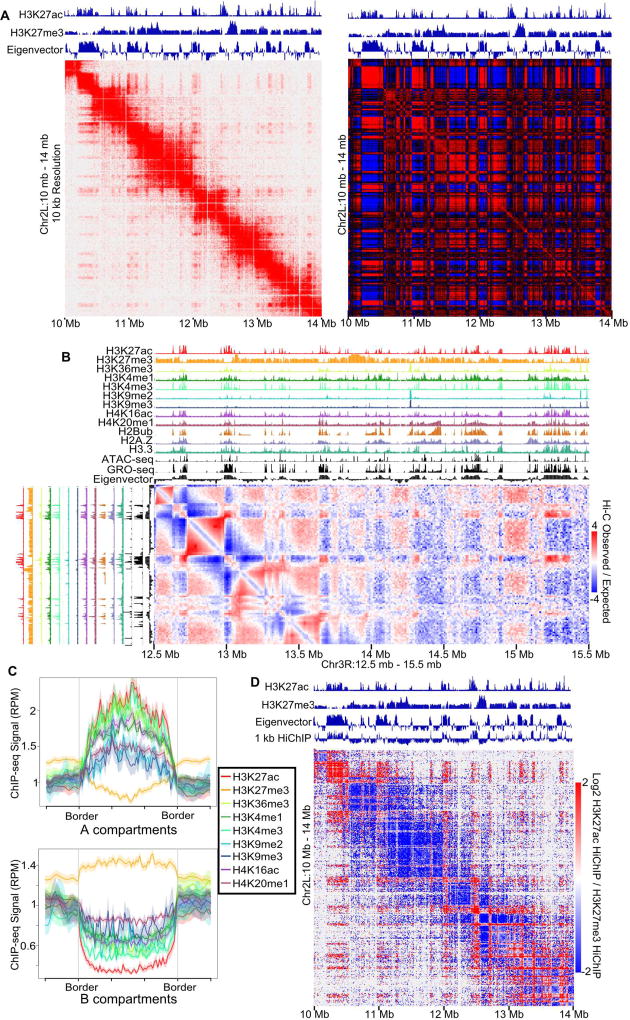
Figure 1. Drosophila has Fine-Scale Compartments.
A. Left: Normalized Hi-C map of Kc167 cells at 10 kb resolution. Right: Pearson Correlation matrix of Hi-C. The eigenvector and H3K27ac and H3K27me3 ChIP-seq are above the Hi-C plot.
B. ChIP-seq for 12 different histone modifications, ATAC-seq, and GRO-seq compared to the Hi-C eigenvector. A slice of the distance normalized Hi-C matrix (observed/expected) is shown corresponding to Chr3R:12.5 Mb – 15.5 Mb (horizontal) and Chr3R:12.5 Mb-13.5 Mb (vertical).
C. Active and inactive chromatin correspond to A and B compartments. Average histone modification profiles over A and B compartments. Color coding of ChIP-seq for histone modifications/variants is indicated.
D. Compartmental interactions defined by HiChIP. Contact map showing differential contacts for H3K27ac vs H3K27me3 HiChIP visualized by Juicebox.
See also Figures S1 and Table S1–S2.