Abstract
Dynamics of human immunodeficiency virus type 1 (HIV-1) tropism after antiretroviral therapy (ART) initiation and their association with disease progression are poorly investigated.
This was a cohort study on subjects from the ICONA cohort receiving ART with persistently detectable (PD) or persistently undetectable (PU) viral load (VL) and with stored plasma or peripheral blood mononuclear cell (PBMC) samples at 2 time-points (T1, T2) after ART initiation. HIV-1 co-receptor tropism was determined by V3-loop sequencing and the geno2pheno algorithm. A switch in viral tropism was defined if the tropism classification at T2 differed from that observed at T1. Time to disease progression, defined as the occurrence of a new acquired immune deficiency syndrome (AIDS)-defining event/death from T2, was also evaluated.
One hundred ninety-five patients were analyzed (124 PD, 71 PU). Over a median follow-up of 22.6 (19.8–28.1) months, PD and PU patients showed similar rates (95% confidence interval) of switch to a non-R5 virus [PD: 6.9 (3.7–11.2)/100-person-years of follow-up (PYFU); PU: 8.0 (3.4–14.5)/100-PYFU; P = 0.63] and of switch to a R5 virus [PD: 15.4 (7.3–26.4)/100-PYFU; PU: 8.1 (2.5–16.7)/100-PYFU; P = 0.38]. Switch to non-R5 virus was predicted by nadir CD4+ before T1.
Twenty-two (18%) PD and 4 (6%) PU subjects experienced disease progression (P = 0.02). The risk of disease progression was independently associated with a switch in co-receptor tropism (adjusted hazard ratio = 4.06, 95% CI: 1.20–13.80, P = 0.03) as well as age, AIDS diagnosis, nadir CD4+ before T2, current CD4+, and VL.
Switch of HIV-1 tropism under ART occurs in both directions, with similar rates in subjects with PD or PU VL and it might be predictive of future unfavorable clinical outcome.
Keywords: CCR5, disease progression, FPR, HIV, tropism switch
1. Introduction
Cross-sectional evaluation of human immunodeficiency virus type 1 (HIV-1) co-receptor usage by means of phenotypic assays and/or V3 population genotyping[1] has been intensively studied in untreated primary[2] and chronic HIV-1 infection[3,4] and in patients on virological failure.[5,6] The impact of baseline tropism on virological response and CD4 gains after antiretroviral therapy (ART) initiation has also been investigated in several studies including or not CCR5 inhibitors.[7–9]
The frequency and implications of co-receptor switch in HIV-1 treated subjects unexposed to CCR5 inhibitors are instead poorly understood. Longitudinal analyses have been performed on co-receptor usage before and after ART initiation in subjects on suppressive therapy,[10–15] virological failure,[15–19] or after therapy interruption,[20,21] yielding controversial results. HIV-1 co-receptor usage has been determined longitudinally under ART in paired plasma[16,17] or peripheral blood mononuclear cell (PBMC) samples[10,12–15] in a very limited subset of patients.
There are several studies that have evaluated the relationship between the co-receptor tropism and the risk of clinical disease progression in ART-naïve subjects or after treatment initiation,[9,22–24] on the other hand, whether HIV-1 tropism switch under ART pressure might be associated with the risk of clinical progression has never been previously investigated.
The aim of this study was to determine the rate of HIV-1 tropism switch in subjects under ART both in presence of persistently detectable (PD) or undetectable (PU) viral load (VL). The association between tropism switch and disease progression was also evaluated.
2. Methods
This is a longitudinal cohort study on adult HIV-1 treated subjects enrolled in the ICONA Foundation Study Cohort, with available paired samples of plasma or cells stored at 2 time-points after ART initiation. Briefly, the ICONA Foundation Cohort is a cohort of HIV-infected patients which superseded the original I.CO.N.A. (Italian Cohort of Antiretroviral-Naïve Patients) study, recruiting HIV-positive patients when still ART-naïve. At their enrollment, subjects provide written informed consent to include their clinical and laboratory data in the ICONA database for scientific purposes.
The ICONA database collects the method of VL quantification and the corresponding detection limit; this information was available for the analyses and used to correctly classify as detectable or undetectable the recorded VL values. VL values > 50 copies/mL were classified as detectable; VL values < 50 copies/mL or below the detection limit in use in each center were classified as undetectable.
Two groups of treated HIV-1 subjects were retrospectively identified based on the recorded VL determinations: subjects with PD VL at and between the 2 considered time-points; subjects with PU VL at and between the 2 considered time-points. Patients treated with maraviroc were excluded from this analysis.
In both groups, the first sample was the nearest to the ART initiation (and approximately at least 6 months after starting), while the second sample was taken approximately 2 years after the date of first sample, with a median interval between the 2 samples of 22.9 (20–28.5) months.
Co-receptor tropism was determined on HIV-RNA from plasma samples in PD subjects and on proviral DNA extracted from PBMC samples in PU subjects.
A single polymerase chain reaction (PCR) product per sample was subjected to standard population sequencing. Sequences were analyzed with Seqscape software v2.5 (Applied Biosystems, Foster City, CA) as previously described.[25–27] Nucleotide mixtures were considered if the second highest peak in the electropherogram was >25%. V3 sequences with more than 8 nucleotide mixtures were discarded. The V3 sequences are available on request.
Co-receptor tropism was inferred with the geno2pheno algorithm setting the false positive rate (FPR) at 10% for plasma samples and 20% for PBMC samples,[1] based on previous studies[28,29] showing concordant results in the determination of co-receptor tropism for the same individual whether plasma RNA or proviral DNA was used. Co-receptor tropism for all PD patients was determined only on plasma RNA at either time-point; similarly in PU patients, co-receptor tropism was assessed only on proviral DNA at either time-point. Clonal prediction was employed for classifying sequences. Clinical isolates were classified as R5 if FPR >10% (RNA) or >20% (DNA) and non-R5 if FPR <10% (RNA) or <20% (DNA). Non-R5 tropism included either R5/X4-coreceptors (dual or mixed tropism) and X4-coreceptor.
Baseline for the statistical analysis used to estimate the rate of tropism switch and factors associated with switch was defined as the date of the first sample (T1).
A co-receptor switch (from R5 to non-R5 or from non-R5 to R5) defined an unstable tropism and was considered to occur if the HIV tropism classification at the second time-point (T2) differed from that observed at T1, otherwise tropism was considered as stable (R5 or non-R5 at both time-points).
Disease progression was defined as the occurrence of a new clinical acquired immune deficiency syndrome (AIDS)-defining event or death; time to disease progression was calculated as the time elapsed since T2 up to the first new AIDS-defining event or death or last available clinical follow-up visit (whichever occurred first).
2.1. Statistical analysis
Patients’ main characteristics were described as median (interquartile range, IQR) for continuous variables or proportions for categorical variables. Continuous variables were compared using the Wilcoxon rank sum test or the Kruskal–Wallis test, as appropriate. Differences between proportions were tested by the χ2 or Fisher exact test.
The incidence rate of co-receptor tropism switch was calculated as the number of switches per 100 person-years of follow-up (PYFU); separate estimates were calculated for switch from non-R5 to R5-tropic viruses and from R5 to non-R5-tropic viruses. Rates were reported with the corresponding 95% confidence interval (95% CI).
Univariate and multivariate Poisson regression models were performed to estimate the risk of tropism switch in relation to the subjects’ virological group (PD vs PU) and other baseline characteristics. Separate models were used to identify predictors of the risk of switch to R5-tropic virus and of the risk of switch to non-R5-tropic virus, and estimates were presented as relative risk (RR) with 95% CI; covariates with a P-value ≤0.20 in the univariate analysis were considered for entry into the multivariate models in addition to age, sex, baseline CD4+, and subjects’ virological group (PU vs PD). Collinearity between variables was assessed by means of the variance inflation factors (VIF); the largest VIF for a single covariate was 1.6, indicating that all variables could be included in the model together.
Mixed-effect linear models, with an unstructured covariance matrix, were used to examine CD4+ trend during the T1–T2 interval or from baseline (T1) until disease progression or last available visit, whichever occurred first; estimates of CD4+ trajectories over time (slope ± standard error) were calculated using random intercept and random slope of months of follow-up to model the within-patient errors.
Univariate and multivariate Cox proportional hazard models were calculated to evaluate the influence of co-receptor tropism switch and other covariates on the risk of disease progression (occurrence of a new AIDS-defining event or death since T2); the contribution of each covariate was expressed by the hazard ratio (HR) with the corresponding 95% CI. The multivariate model included demographic variables, hepatitis C virus (HCV) co-infection (a factor with a potential impact on disease progression) and those confounders with a P≤0.10 in the univariate analysis: age, sex, HCV co-infection, AIDS diagnosis before T2, CD4 nadir before T2, years of ART treatment, co-receptor tropism switch, current CD4+, current HIV-RNA.
In all statistical analyses, a P < 0.05 was considered statistically significant. The analyses were performed using SAS Software, release 9.2 (SAS Institute, Cary, NC).
3. Results
3.1. Patients and their characteristics
HIV-1 V3 loop sequences were successfully obtained from 195 subjects (124 PD, 71 PU) satisfying the study inclusion criteria at baseline (T1) and at T2; baseline was 10.0 (5.2–16.3) months after the date of ART initiation.
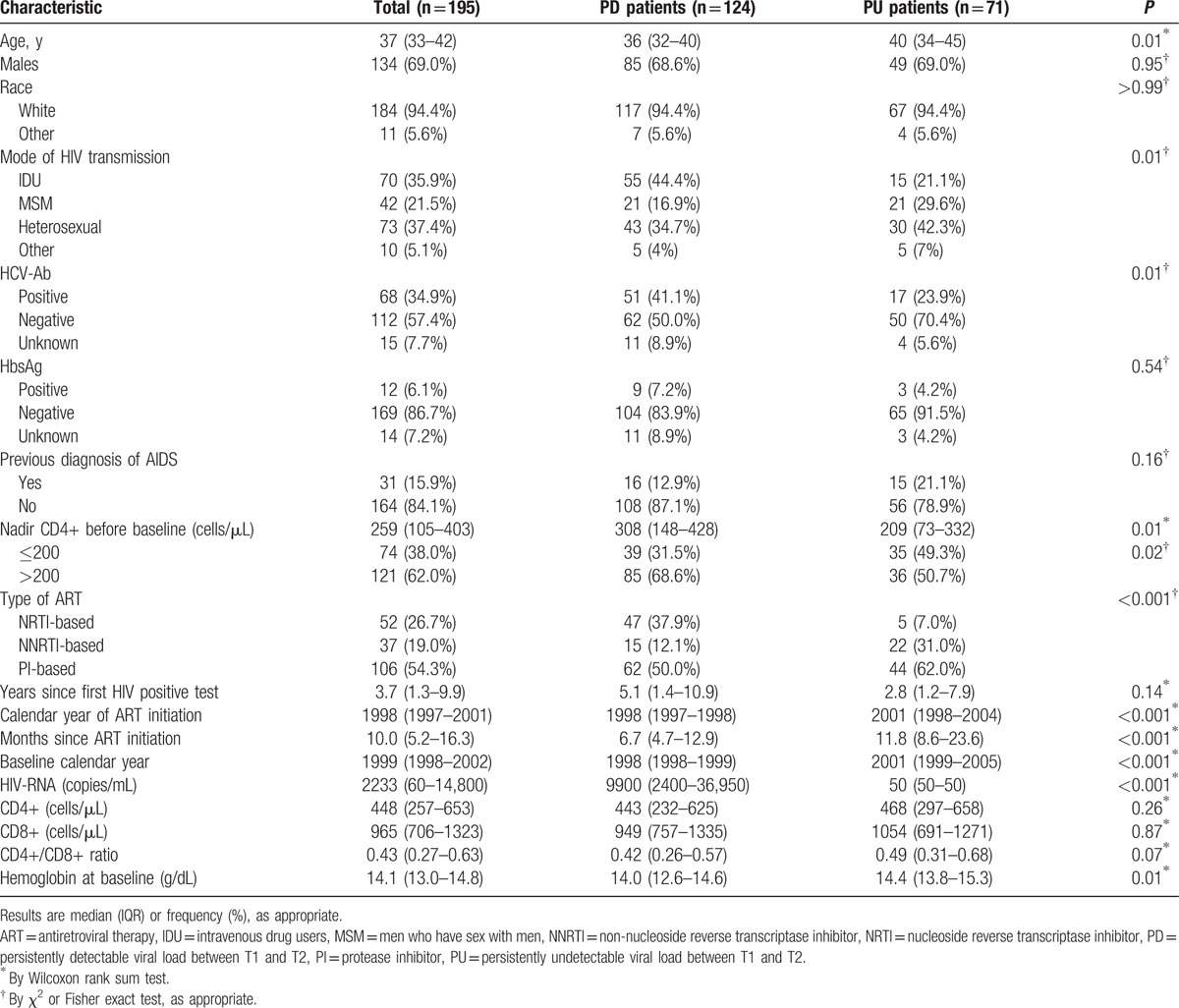
The main baseline characteristics of PD and PU subjects are shown in Table 1; PD subjects were younger, more frequently intravenous drug users, more likely to be co-infected with HCV (41.1% vs 23.9%, P = 0.01), with an earlier calendar year of ART initiation, more frequently treated with a nucleoside reverse transcriptase inhibitor (NRTI)-based regimen (37.9% vs 7.0%, P < .001), as compared with PU patients. The median number of VL determinations between the 2 time-points was 6 (5–8) and 8 (6–10) in PD and PU subjects, respectively (P < .001). In PD subjects, HIV-RNA area-under-the-curve between the 2 time-points was 3.82 (3.31–4.26) log10 copies/mL/d. Overall, subtype B HIV-1 was assigned to 183 patients [117/124 (94%) in PD and 66/71 (93%) in PU, P = 0.76].
Table 1.
Baseline demographic, clinical, and laboratory characteristics of 195 HIV-1 treated subjects included in the analysis.
3.2. HIV-1 tropism switch
At baseline (T1), 93 (75%) PD patients were predicted to harbor an R5 virus in comparison to 46 PU (65%) subjects, with no significant difference (P = 0.14). At T2, there were 90 (73%) PD subjects with an R5 virus after a median follow-up of 22.1 (19.2–24.7) months and 43 (61%) PU subjects had an R5 strain after a median follow-up of 24.4 (21.0–31.6) months (difference not statistically significant: P = 0.11).
There were 101 (81%) PD and 58 (82%) PU subjects with stable co-receptor tropism and 23 (19%) PD and 13 (18%) PU subjects experienced a co-receptor tropism switch between the 2 time-points (Fig. 1). The rate of co-receptor switch was similar between PD and PU subjects (Fig. 1). Among PD subjects, the switch was mainly from a non-R5 to an R5 strain (P = 0.04) while no difference was found among PU subjects (P = 0.81).
Figure 1.
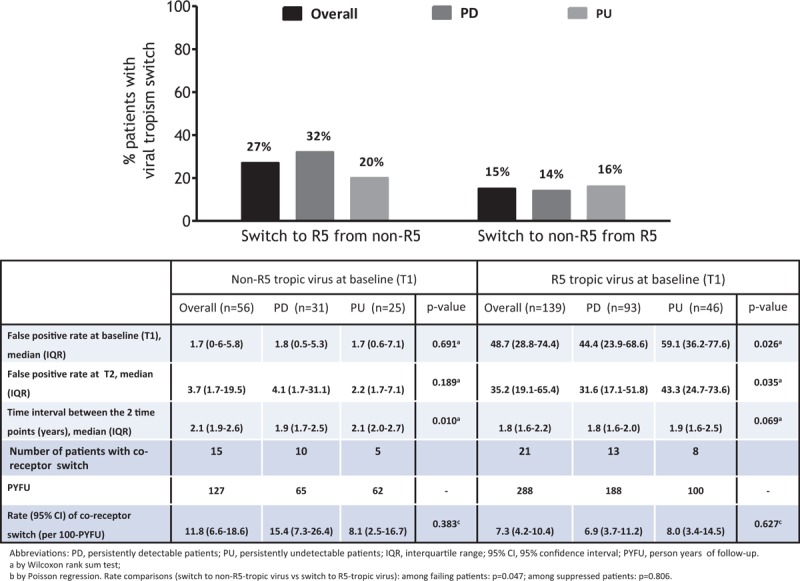
Co-receptor tropism switch in subjects receiving antiretroviral therapy (ART) with persistently detectable (PD) or persistently undetectable (PU) viral load.
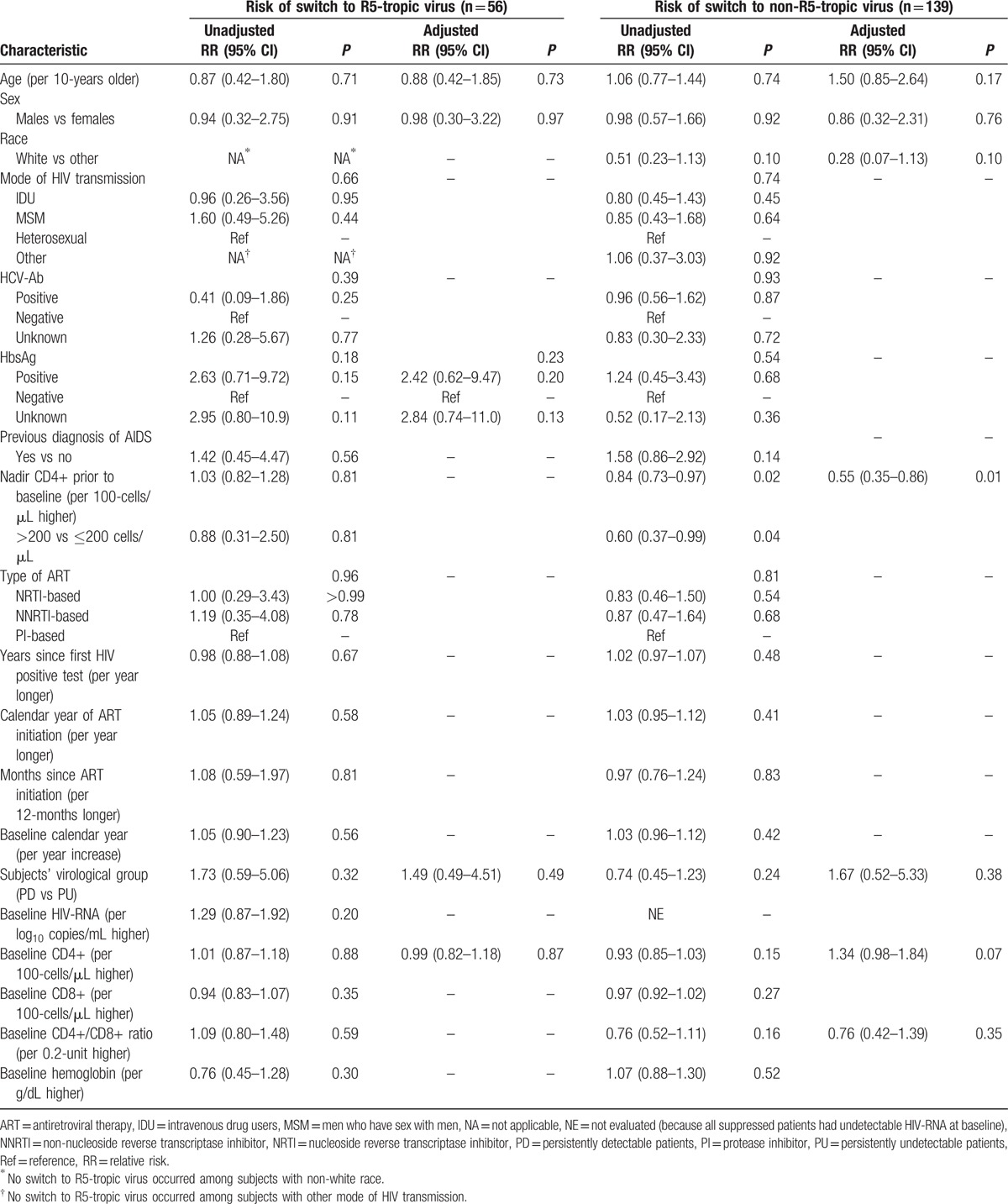
No association was found between all the considered baseline demographic, clinical, and laboratory characteristics or the subjects’ group (i.e., being PD or PU) with the risk of tropism switch to R5, while nadir CD4+ before T1 was the only factor associated with the risk of tropism switch to non-R5 [adjusted RR (per 100-cells/μL higher) = 0.55, 95% CI: 0.35–0.86, P = 0.01] (Table 2).
Table 2.
Poisson regression: unadjusted and adjusted relative risk of co-receptor switch in the 195 HIV-1 treated subjects considered in the analysis.
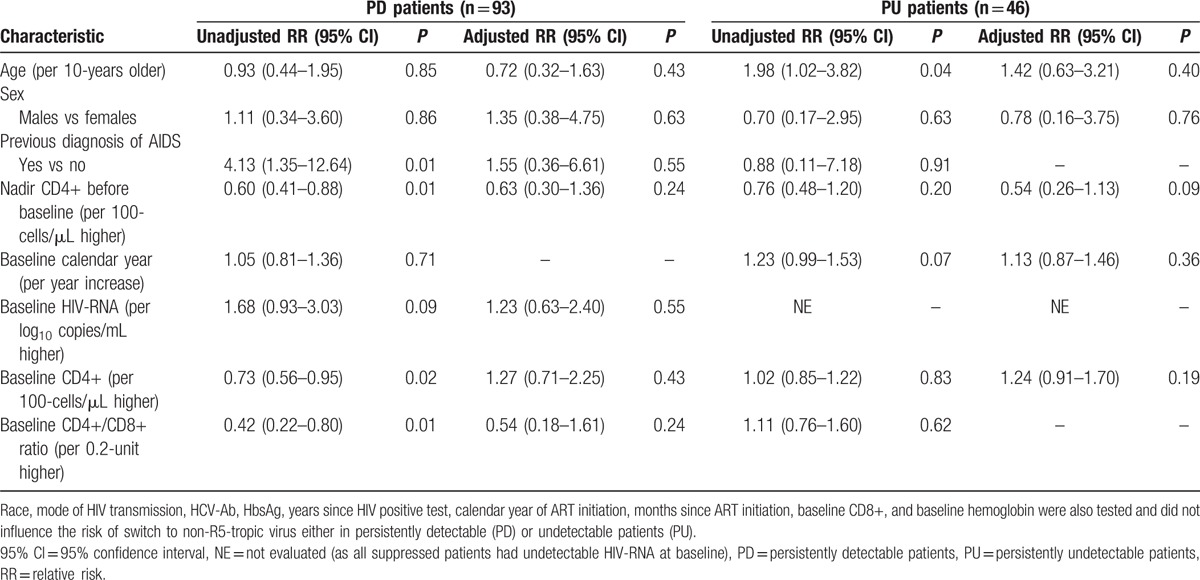
When PD and PU patients were considered separately, no association was observed between all the considered factors and the risk of tropism switch to R5-tropic virus (data not shown) or the risk of switch to non R5-tropic virus (Table 3).
Table 3.
Poisson regression: unadjusted and adjusted relative risk of switch to non-R5-tropic virus in the 139 HIV-1 treated subjects with an R5-tropic virus at baseline.
We explored the association between co-receptor switch and changes in CD4+ and VL during the T1–T2 time interval (Table 4): a less favorable CD4+ trend was observed in subjects with tropism switch in comparison to patients with stable tropism. Results were similar when restricting to the switch from a non-R5 to an R5 strain or vice versa.
Table 4.
Immuno-virological trend between the 2 time points according to co-receptor switch and patients’ virological group.

3.3. Association between co-receptor switch and HIV-1 disease progression
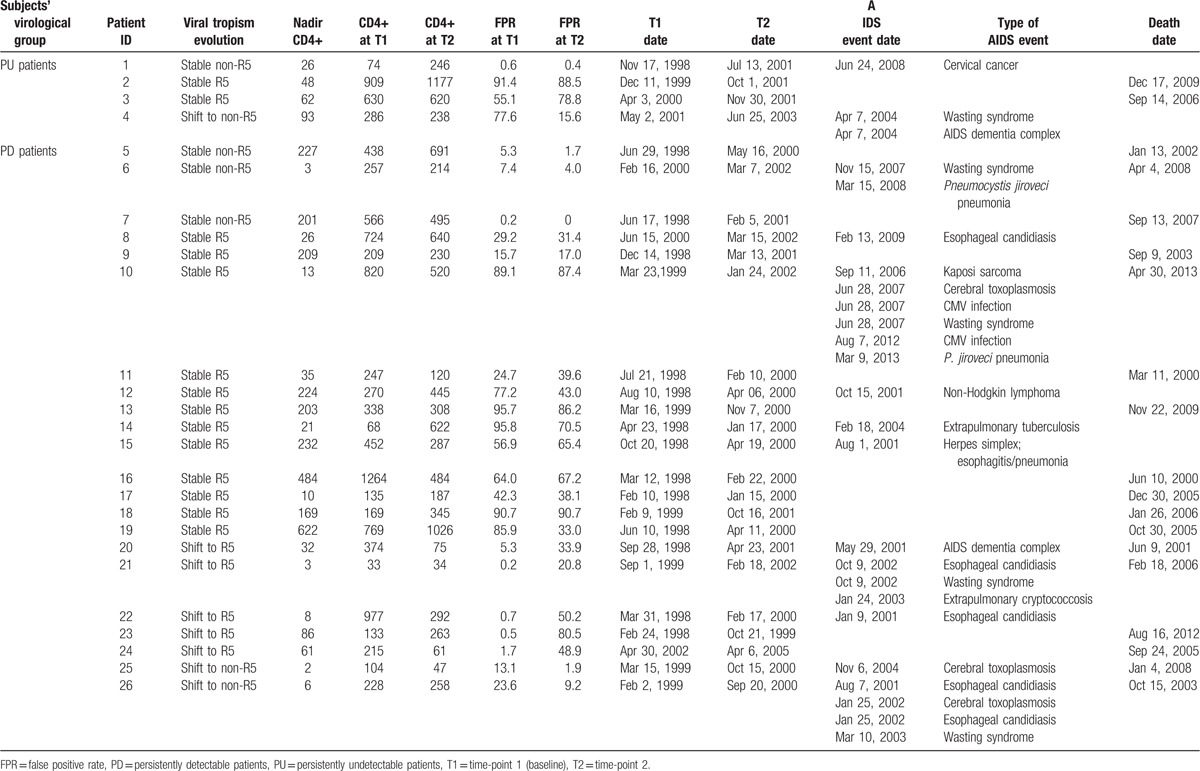
After a median time of 5.4 (2.5–9.3) years from T2, 26 (13%) patients developed at least one AIDS event or died: 22/124 (18%) among the PD group and 4/71 (6%) among PU subjects, respectively (P = 0.02). Among the 22 PD subjects, 11 patients had at least one AIDS event and 17 patients died (11 without AIDS events); among PU subjects, 2 patients had new AIDS events, 2 patients died. Clinical details of PD and PU subjects with disease progression are reported in Table 5.
Table 5.
Characteristics of 26 patients with disease progression from T2.
PD subjects showed a faster disease progression and were associated with a higher risk of disease progression as compared with PU subjects (unadjusted HR = 3.09, 95% CI: 1.06–9.00, P = 0.04].
Patients with tropism switch appeared to have faster disease progression rather than those with stable R5 or non-R5 co-receptor tropism and the risk of disease progression was higher among subjects with tropism switch (to R5 or non-R5) in comparison to those with stable tropism (R5 or non-R5) (HR = 2.39, 95% CI: 1.03–5.54, P = 0.04].
At multivariate analysis, the risk of disease progression was more likely in older subjects, in subjects with an AIDS diagnosis before T2, with a lower CD4+ count nadir before T2, with lower current CD4+, with higher current VL, and in subjects with co-receptor switch rather than those with stable tropism (Table 6).
Table 6.
Cox proportional hazard models: unadjusted and adjusted relative risk of HIV disease progression in the 195 HIV-1 treated subjects.

4. Discussion
The dynamics of co-receptor tropism switch in HIV-1 infected subjects under ART pressure remain poorly explored. Our findings show that in approximately 2 years of follow-up under ART, co-receptor usage is stable in 80% of cases, either in case of successful control of viral replication or in case of persistent detectable HIV-1 viremia. Switch of HIV-1 tropism under ART occurs in both directions, with similar rates in subjects with PD or PU VL. Although comparisons with previous studies are difficult because of differences in study design, length of follow-up, methodologies, and cutoffs used for co-receptor determination, our data confirmed on a large-scale findings reported in a small case series. Switch from non-R5 to R5 virus has been described in detail in a previous study evaluating the co-receptor usage in 40 treated failing patients with a median interval between the 2 time-points of about 20 months.[16] Co-receptor usage was stable in 87% of patients, conversion from X4 to R5 or from R5 to X4 was observed in 2 patients, respectively. In another study[19] on 76 subjects with drug-resistant viremia >1000 copies/mL on a stable ART regimen for >4 months and with a median of 3 tropism determinations (by means of a phenotypic recombinant virus assay) over 9 months, the rate of tropism switch was about 10% and occurred in both directions; 12% of patients infected with R5-tropic virus at baseline switched to D/M-tropic virus by 1 year and 11% infected with D/M-tropic virus at baseline switched to R5 or X4-tropic virus over the same period. In this study, the apparent tropism switches were considered as oscillations in the amount of X4-tropic virus around the limit of assay detection. Nevertheless, a possible alternative interpretation might be that the switch of the co-receptor usage from X4 to R5 under ART pressure may indeed truly occur, independently from CD4 level and undetectability of VL. In fact, a tropism switch from non-R5 to R5 virus has also been observed in a very small series of patients on suppressive ART[10–15] or interrupting successful ART.[20,21]
Both R5 and non-R5 variants have been shown to persist in viral reservoirs after prolonged suppression of plasma VL below the detection limit.[10–15] Preliminary studies documented that the stability of CCR5 co-receptor usage is correlated with the maintenance of HIV-RNA <2.5 copies/mL.[12] Correlation analyses between stability of the co-receptor usage and residual viremia, changes in proviral DNA and viral replicative capacity[30] will provide new insights into the mechanisms of co-receptor tropism switching in subjects with virological suppression.
We were not able to identify predictors of tropism switch with the only exception of CD4 nadir that was predictive of the switch from R5 to non-R5 which might be along the lines of previous studies showing that the presence of non-R5 virus is associated with low nadir CD4 values.[31,32]
Results of the impact of tropism switch on long-term risk of disease progression in subjects receiving ART are lacking. Previous studies evaluated the risk of disease progression in relation to co-receptor tropism by geno2pheno algorithm determined at a single time-point yielding conflicting results.[9,33]
In our analysis, the presence of unstable co-receptor tropism, irrespectively of tropism switch direction, was associated with less favorable CD4 changes and with an increased risk of disease progression, especially in subjects with PD VL. Our results might contribute to explain why a substantial number of patients (62%) diagnosed at a late stage of HIV-1 infection had only R5-tropic virus strains.[4]
Main limitations of our study included the small number of individuals experiencing the study outcomes and, consequently, the power to detect predictive factors of co-receptor tropism switch or the disease progression risk and the possibility to evaluate if the direction of tropism switch may differently affect the risk of disease progression. In addition, we cannot exclude that some individuals with stable tropism might also have experienced tropism switch before ART initiation, between ART initiation and T1, or between T2 and the date of disease progression.
In conclusion, HIV-1 tropism switch under ART pressure was detected in both directions and it appears to occur with similar rates in subjects with PD or PU VL. An unstable tropism might be associated with a less favorable CD4 change and an increased risk of disease progression, independently of established risk factors for HIV progression. Larger studies are needed to confirm our hypothesis.
Acknowledgments
ICONA Foundation Study Group - Board of Directors: A. d’Arminio Monforte (Vice-President), M. Andreoni, G. Angarano, A. Antinori, F. Castelli, R. Cauda, G. Di Perri, M. Galli, R. Iardino, G. Ippolito, A. Lazzarin, C. F. Perno, F. von Schloesser, P. Viale.
Scientific Secretary: A. d’Arminio Monforte, A. Antinori, A. Castagna, F. Ceccherini-Silberstein, A. Cozzi-Lepri, E. Girardi, S. Lo Caputo, C. Mussini, M. Puoti.
Steering Committee: M. Andreoni, A. Ammassari, A. Antinori, C. Balotta, P. Bonfanti, S. Bonora, M. Borderi, M. R. Capobianchi, A. Castagna, F. Ceccherini-Silberstein, A. Cingolani, P. Cinque, A. Cozzi-Lepri, A. d’Arminio Monforte, A. De Luca, A. Di Biagio, E. Girardi, N. Gianotti, A. Gori, G. Guaraldi, G. Lapadula, M. Lichtner, S. Lo Caputo, G. Madeddu, F. Maggiolo, G. Marchetti, S. Marcotullio, L. Monno, C. Mussini, M. Puoti, E. Quiros Roldan, S. Rusconi, A. Saracino.
Statistical and Monitoring Team: A. Cozzi-Lepri, I. Fanti, L. Galli, P. Lorenzini, A. Rodano, M. Shanyinda, A. Tavelli.
Participating Physicians and Centers: Italy A. Giacometti, A. Costantini, S. Mazzoccato (Ancona); G. Angarano, L. Monno, C. Santoro (Bari); F. Maggiolo, C. Suardi (Bergamo); P. Viale, E. Vanino, G. Verucchi (Bologna); F. Castelli, E. Quiros Roldan, C. Minardi (Brescia); T. Quirino, C. Abeli (Busto Arsizio); P. E. Manconi, P. Piano (Cagliari); J. Vecchiet, K. Falasca (Chieti); L. Sighinolfi, D. Segala (Ferrara); F. Mazzotta, S. Lo Caputo (Firenze); G. Cassola, C. Viscoli, A. Alessandrini, R. Piscopo, G. Mazzarello (Genova); C. Mastroianni, V. Belvisi (Latina); P. Bonfanti, I. Caramma (Lecco); A. Chiodera, A. P. Castelli (Macerata); M. Galli, A. Lazzarin, G. Rizzardini, M. Puoti, A. d’Arminio Monforte, A. L. Ridolfo, R. Piolini, A. Castagna, S. Salpietro, L. Carenzi, M. C. Moioli, C. Tincati, G. Marchetti (Milano); C. Mussini, C. Puzzolante (Modena); A. Gori, G. Lapadula (Monza); N. Abrescia, A. Chirianni, G. Borgia, M. G. Guida, M. Gargiulo, I. Gentile, R. Orlando (Napoli); F. Baldelli, D. Francisci (Perugia); G. Parruti, T. Ursini (Pescara); G. Magnani, M. A. Ursitti (Reggio Emilia); R. Cauda, M. Andreoni, A. Antinori, V. Vullo, A. Cingolani, A. d’Avino, L. Gallo, E. Nicastri, R. Acinapura, M. Capozzi, R. Libertone, G. Tebano, M. Zaccarelli (Roma); F. Viviani, L. Sasset (Rovigo); M. S. Mura, G. Madeddu (Sassari); A. De Luca, B. Rossetti (Siena); P. Caramello, G. Di Perri, G. C. Orofino, S. Bonora, M. Sciandra (Torino); M. Bassetti, A. Londero (Udine); G. Pellizzer, V. Manfrin (Vicenza).
Footnotes
Abbreviations: 95% CI = 95% confidence interval, AIDS = acquired immune deficiency syndrome, ART = antiretroviral therapy, FPR = false positive rate, HCV = hepatitis C virus, HIV-1 = human immunodeficiency virus type 1, HR = hazard ratio, IQR = interquartile range, NNRTI = non-nucleoside reverse transcriptase inhibitor, NRTI = nucleoside reverse transcriptase inhibitor, PBMC = peripheral blood mononuclear cell, PD = persistently detectable (viral load), PU = persistently undetectable (viral load), PYFU = person-years of follow-up, RR = relative risk, T1 = time-point 1, T2 = time-point 2, VIF = variance inflation factor, VL = viral load.
This study was presented in part at the 14th European AIDS Conference, Brussels, Belgium, October 16–19, 2013.
Funding/support: This work was supported by ViiV Healthcare, grant no. 3000080700, and by the European Commission Framework 7 Programme (CHAIN, the Collaborative HIV and Anti-HIV Drug Resistance Network, Integrated Project no. 223131).
AC has received consultancy payments and speaking fee from Bristol-Myers Squibb, Gilead, ViiV Healthcare, Merck Sharp & Dohme, and Janssen-Cilag. ADL has received consultancy payments and speaking fee from Bristol-Myers Squibb, Gilead Sciences, ViiV Healthcare, Merck Sharp & Dohme, ABBvie, Janssen-Cilag, and Teva Pharmaceuticals. FC-S has received consultancy payments and speaking fee from Bristol-Myers Squibb, Gilead Sciences, ViiV Healthcare, Merck Sharp & Dohme, ABBvie, Janssen-Cilag, and Roche Diagnostics. ADM has received consultancy payments and speaking fee from Bristol-Myers Squibb, Gilead Sciences, ViiV Healthcare, Merck Sharp & Dohme, ABBvie, and Janssen-Cilag. The remaining authors have no conflicts of interest to disclose.
References
- 1.Vandekerckhove LP, Wensing AM, Kaiser R, et al. European guidelines on the clinical management of HIV-1 tropism testing. Lancet Infect Dis 2011; 11:394–407. [DOI] [PubMed] [Google Scholar]
- 2.Rieder P, Joos B, Scherrer AU, et al. Characterization of human Immunodeficiency virus Type 1 (HIV-1) diversity and tropism in 145 patients with primary HIV-1 infection. Clin Infect Dis 2011; 53:1271–1279. [DOI] [PubMed] [Google Scholar]
- 3.Simon B, Grabmeier-Pfistershammer K, Rieger A, et al. HIV coreceptor tropism in antiretroviral treatment-naïve patients newly diagnosed at a late stage of HIV infection. AIDS 2010; 24:2051–2058. [DOI] [PubMed] [Google Scholar]
- 4.Soulie C, Charpentier C, Flandre C, et al. Natural evolution of CD4+ Cell Count in patients with CD4 > 350 or > 500 cells/mmc at the time of diagnosis according to HIV-1 coreceptor tropism. J Med Virol 2012; 84:1853–1856. [DOI] [PubMed] [Google Scholar]
- 5.Hunt PW, Harrigan R, Huang W, et al. Prevalence of CXCR4 tropism among antiretroviral-treated HIV-1-infected patients with detectable viremia. J Infect Dis 2006; 194:926–930. [DOI] [PubMed] [Google Scholar]
- 6.Ferrer P, Tello B, Montecinos L, et al. Prevalence of R5 and X4 variants in antiretroviral experienced patients with virological failure. J Clin Virol 2014; 60:290–294. [DOI] [PubMed] [Google Scholar]
- 7.Seclen E, Soriano V, Gonzales M, et al. Impact of baseline HIV-1 tropism on viral response and CD4 cell count gains in HIV-Infected patients receiving first-line antiretroviral therapy. J Infect Dis 2011; 204:139–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilkin JW, Goetz MB, Leduc R, et al. Reanalysis of coreceptor tropism in HIV-1 infected adults using a phenotypic assay with enhanced sensitivity. Clin Infect Dis 2011; 52:925–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waters L, Mandalia S, Randell P, et al. The impact of HIV tropism on decrease in CD4 cell count, clinical progression, and subsequent response to first antiretroviral therapy regimen. Clin Infect Dis 2008; 46:1617–1623. [DOI] [PubMed] [Google Scholar]
- 10.Saracino A, Monno L, Cibelli DC, et al. Co-receptor switch during CART is independent of virological success. J Med Virol 2009; 81:2036–2044. [DOI] [PubMed] [Google Scholar]
- 11.Soulie C, Lambert-Niclot S, Wirden M, et al. Low frequency of HIV-1 tropism evolution in patients successfully treated for at least 2 years. AIDS 2011; 25:537–539. [DOI] [PubMed] [Google Scholar]
- 12.Parisi SG, Andreis S, Mengoli G, et al. A stable CC-chemokine receptor (CCR)-5 tropic virus is correlated with the persistence of HIV RNA at less than 2.5 copies in successfully treated naïve subjects. BMC Infect Dis 2013; 13:314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raymond S, Saliou A, Delobel P, et al. Evolution of HIV-1 quasispecies and coreceptor use in cell reservoirs of patients on suppressive antiretroviral therapy. J Antimicrob Chemother 2014; 69:2527–2530. [DOI] [PubMed] [Google Scholar]
- 14.Meini G, Rossetti B, Bianco C, et al. Longitudinal analysis of HIV-1 coreceptor tropism by single and triplicate HIV-RNA and DNA sequencing in patients undergoing successful first-line antiretroviral therapy. J Antimicrob Chemother 2014; 69:735–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rozera G, Abbate I, Giombini E, et al. Evolution of HIV-1 tropism at quasispecies level after 5 years of combination antiretroviral therapy in patients always suppressed or experiencing episodes of virological failure. J Antimicrob Chemother 2014; 69:3085–3094. [DOI] [PubMed] [Google Scholar]
- 16.Lehman C, Daumer M, Boussaad I, et al. Stable coreceptor usage of HIV in patients with ongoing treatment failure on HAART. J Clin Virol 2006; 37:300–304. [DOI] [PubMed] [Google Scholar]
- 17.Briz V, Poveda E, del Mar Gonzaléz M, et al. Impact of antiretroviral therapy on viral tropism in HIV-infected patients followed longitudinally for over 5 years. J Antimicrob Chemother 2008; 61:405–410. [DOI] [PubMed] [Google Scholar]
- 18.Lee GQ, Dong W, Mo T, et al. Limited evolution of inferred HIV-1 tropism while viremia is undetectable during standard HAART therapy. PLoS One 2014; 9:e99000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunt P, Huang W, Coakley E, et al. Longitudinal evaluation of viral co-receptor tropism switches among HIV-Infected patients with drug resistant viremia. 14th Conference on Retroviruses and Opportunistic Infections, Los Angeles, February 25–28 2007; abstract 619. [Google Scholar]
- 20.Sarmati L, Parisi S G, Andreoni C, et al. Switching of inferred tropism caused by HIV during Interruption of antiretroviral therapy. J Clin Microbiol 2010; 48:2586–2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waters LJ, Scourfield AT, Marcano M, et al. The evolution of coreceptor tropism in HIV-infected patients interrupting suppressive antiretroviral therapy. Clin Infect Dis 2011; 52:671–673. [DOI] [PubMed] [Google Scholar]
- 22.Daar ES, Kesler KL, Petropoulos CJ, et al. Baseline HIV type 1 coreceptor tropism predicts disease progression. Clin Infect Dis 2007; 45:643–649. [DOI] [PubMed] [Google Scholar]
- 23.Goetz MB, Leduc R, Kostman JR, et al. Relationship between HIV coreceptor tropism and disease progression in persons with untreated chronic HIV infection. J Acquir Immune Defic Syndr 2009; 50:259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weiser B, Philpott S, Klimkait T, et al. HIV coreceptor usage and CXCR4- specific viral load predict clinical diseases progression during combination antiretroviral therapy. AIDS 2008; 22:469–479. [DOI] [PubMed] [Google Scholar]
- 25.Svicher V, D’Arrigo R, Alteri C, et al. OSCAR Study Group. Performance of genotypic tropism testing in clinical practice using the enhanced sensitivity version of Trofile as reference assay: results from the OSCAR Study Group. New Microbiol 2010; 33:195–206. [PubMed] [Google Scholar]
- 26.Svicher V, Alteri C, Montano M, et al. Genotypic testing on HIV-1 DNA as a tool to assess HIV-1 co-receptor usage in clinical practice: results from the DIVA study group. Infection 2014; 42:61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monno L, Saracino A, Scudeller L, et al. Impact of mutations outside the V3 region on coreceptor tropism phenotypically assessed in patients infected with HIV-1 subtype B. Antimicrob. Agents Chemother 2011; 55:5078–5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baumann RE, Rogers AA, Hamdan HB, et al. Determination of HIV-1 coreceptor tropism using proviral DNA in women before and after viral suppression. AIDS Res Ther 2015; 12:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verhofstede C, Brudney D, Reynaerts J, et al. Concordance between HIV-1 genotypic coreceptor tropism predictions based on plasma RNA and proviral DNA. HIV Med 2011; 12:544–552. [DOI] [PubMed] [Google Scholar]
- 30.Claiborne DT, Prince JL, Scully E, et al. Replicative fitness of HIV-1 drives acute immune activation, proviral load in memory CD4+ T cells and disease progression. Proc Natl Acad Sci U S A 2015; 112:E1480–E1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hunt PW, Harrigan PR, Huang W, et al. Prevalence of CXCR4 tropism among antiretroviral-treated HIV-1-infcted patients with detectable viremia. J Infect Dis 2006; 194:926–930. [DOI] [PubMed] [Google Scholar]
- 32.Soulié C, Fourati S, Lambert-Niclot S, et al. Factors associated with proviral DNA HIV-1 tropism in antiretroviral therapy-treated patients with fully suppressed plasma HIV viral load: implications for the clinical use of CCR5 antagonists. J Antimicrob Chemother 2010; 65:749–751. [DOI] [PubMed] [Google Scholar]
- 33.Fontdevila MC, Cozzi-Lepri A, Phillips A, et al. Plasma HIV-tropism and risk of short-term clinical progression to AIDS or death. International Congress of Drug Therapy in HIV Infection, Glasgow, UK, November 2–6 2014; abstract P158. [Google Scholar]


