Fig. 2.
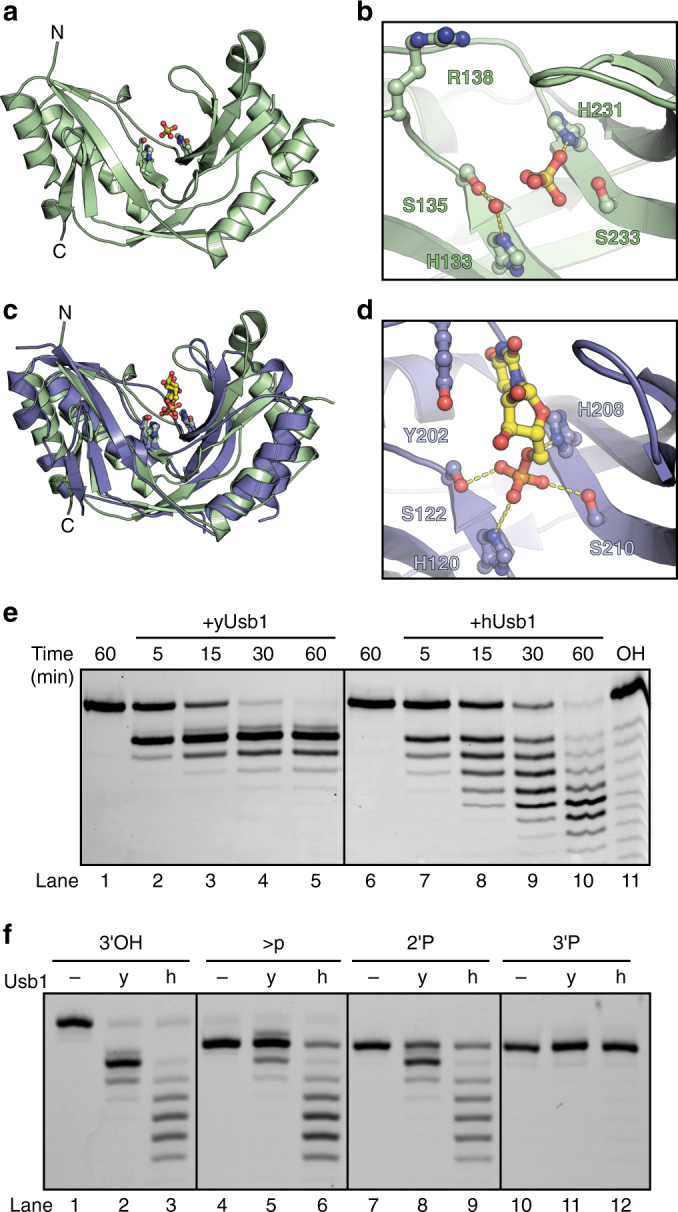
Structure of yUsb1 and structure of hUsb1 with a substrate analog bound in the active site. a Structure of yUsb1 at 1.8 Å. Residues 1–70 were excluded from the crystallizable construct as they were predicted to be unstructured. The two active site H-X-S motifs are shown in ball-and-stick representation. b Active site of yUsb1. A sulfate ion is coordinated in the center of the H-X-S motifs. A water molecule is coordinated by histidine 133 and serine 135. (c) Structure of hUsb1 with 5′ UMP in the active site (blue; 5′ UMP in yellow) is similar to the structure of yUsb1 (green). Residues 1–78 were truncated to facilitate crystallization. The two active site H-X-S motifs are shown in sticks. d Active site of hUsb1. 5′ UMP is coordinated in the center of the H-X-S motifs. The 5′ phosphate is positioned similarly to the sulfate ion in the yUsb1 structure (b). Tyrosine 202 makes a stacking interaction with the uracil of 5′ UMP. Hydrogen bonds between histidine 208 and the O5′ oxygen and between serine 122, histidine 120 and serine 210 and the phosphate hold 5′ UMP in the active site. e Time course comparing yUsb1 (lanes 1–5) and hUsb1 (lanes 6–10) activity. An alkaline hydrolysis ladder (lane 11) shows the mobility of oligonucleotide products of different length. f Single time-point (60 min) comparison of yUsb1 (y) and hUsb1 (h) on RNA substrates with different 3′ modifications. Both yUsb1 and hUsb1 are most active on 2′,3′-cis-diol RNAs (lanes 1–3), less active on cyclic phosphate (lanes 4–6) and 2′ phosphate (lanes 7–9) RNAs and inactive on 3′ phosphate RNAs
