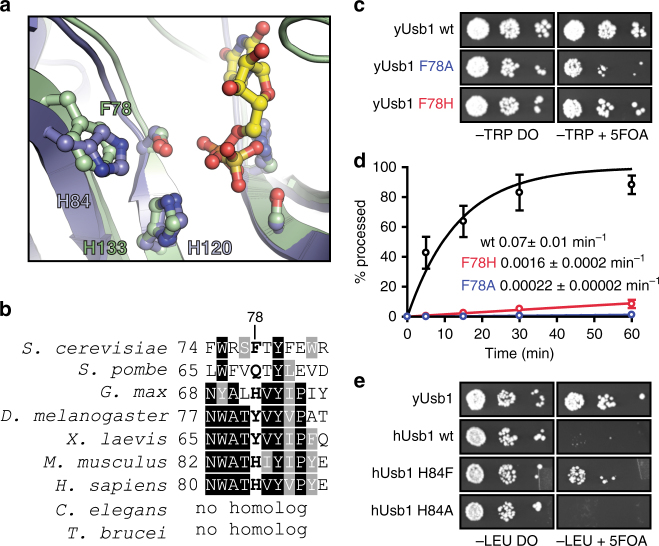
Fig. 3.
Residue F78 influences RNA processing by yUsb1. a Overlay of yUsb1 and hUsb1 active sites shows that yUsb1-F78 and hUsb1-H84 are positioned similarly adjacent to the active site. b Sequence alignment of the region surrounding yUsb1-F78/hUsb1-H84. Several residues (yUsb1 W75 and Y80) are perfectly or near-perfectly conserved, while conservation of position F78 is limited to aromaticity. c Usb1 mutants F78A and F78H can complement genomic deletion of USB1 using plasmid shuffle and assaying for growth on media containing 5-fluoroorotic acid (5-FOA). Usb1-F78A results in a growth defect. d Quantification of the rate of Usb1 processing on a fluorescent substrate (as in Fig. 1) with different substitutions at position 78. Plotted data points represent the average of three technical replicates ± s.d. e Overexpression of yUsb1 (under control of the GPD promoter) complements deletion of USB1, but overexpression of hUsb1 results in an extreme slow growth phenotype. Overexpression of hUsb1-H84F partially rescues growth, while hUsb1-H84A does not