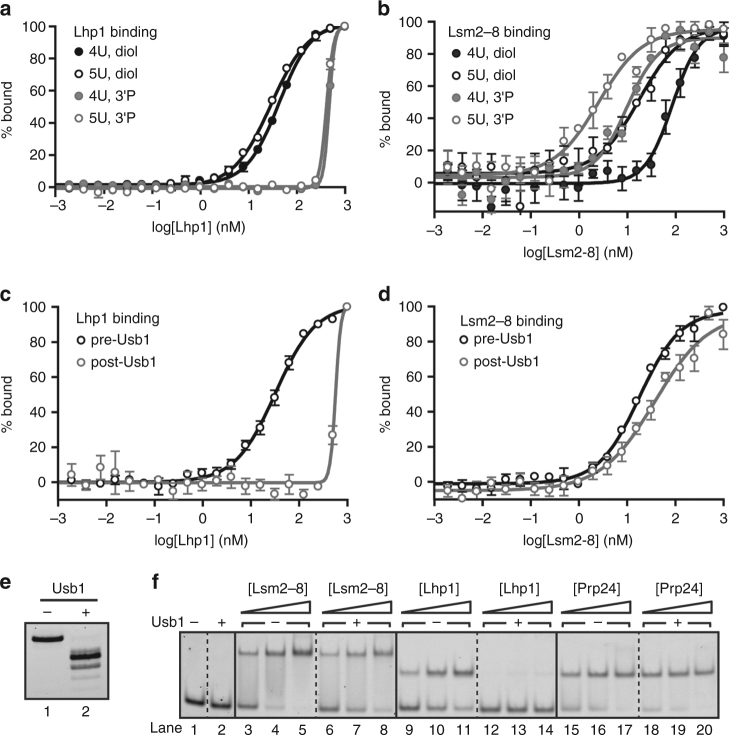
Fig. 5.
Usb1 processing influences RNP formation. a Fluorescence polarization binding data comparing Lhp1 binding to U6 95-112 with a cis-diol (black, filled circles), U6 95-112+1U with a cis-diol (black, open circles), U6 95–112 with a 3′ phosphate (gray, filled circles), and U6 95–112+1U with a 3′ phosphate (gray, open circles). Plotted data points represent the average of three technical replicates ± s.d. for (a–d). b Fluorescence polarization binding data comparing Lsm2–8 binding to U6 95–112 with a cis-diol (black, filled circles), U6 95–112+1U with a cis-diol (black, open circles), U6 95–112 with a 3′ phosphate (gray, filled circles) and U6 95–112+1U with a 3′ phosphate (gray, open circles). c Fluorescence polarization binding data comparing Lhp1 binding to U6 95–112+1U before (black) and after (gray) Usb1 processing. d Fluorescence polarization binding data comparing Lsm2–8 binding to U6 95–112+1U before (black) and after (gray) Usb1 processing. e Denaturing gel comparing RNAs used in c, d. f The affinities of Lsm2–8 and Lhp1 for full-length U6 are influenced by Usb1 processing. Native gel analysis comparing Lsm2–8, Lhp1 and Prp24 affinity for U6 RNA before and after treatment with Usb1. Usb1 processing does not change the mobility of U6 on a native gel (lanes 1 vs. 2). Lsm2–8 binds similarly before (lanes 3–5) and after (lanes 6–8) Usb1 processing. Lhp1 binding (lanes 9–11) is negligible after Usb1 processing (lanes 12–14). Prp24 binding is unchanged before (lanes 15–17) and after Usb1 processing (lanes 18–20)