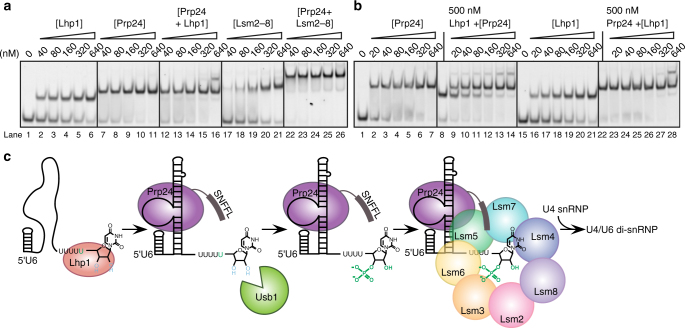
Fig. 6.
The U6 snRNP assembly pathway. a Native gel analysis of U6 binding partners. Lhp1 and Prp24 bind U6 1–112 with a cis-diol tightly (lanes 2–6 and 7–11). Inclusion of equimolar amounts of Lhp1 and Prp24 does not promote formation of a ternary complex except at the highest concentration (lanes 12–16). In contrast, Lsm2–8 binds U6 relatively weakly (17–21), but upon inclusion of Prp24 (lanes 22–26), Lsm2–8 efficiently forms a co-complex of U6, Lsm2–8 and Prp24. b Prp24 binds naked U6 1–112 with a cis-diol (lanes 2–7) and U6 pre-saturated withLhp1 (lanes 9–14) tightly. Prp24 abstracts U6 from U6-Lhp1 much more efficiently than it forms a U6/Prp24/Lhp1 complex. Lhp1 binds naked U6 (lane 21), but cannot bind or release U6 from pre-formed U6-Prp24. c Model of U6 snRNP assembly. U6 is synthesized by RNA polymerase III and initially bound by Lhp1. Binding of Prp24 weakens Lhp1 affinity for the 3′ tail of U6, allowing Usb1 to remove a uridine and leave a 3′ phosphate modified tail. Lsm2–8 recognizes the 3′ tail of U6 and interacts with Prp24 to form the U6 snRNP, which can then be assembled into the spliceosome via the U4/U6 di-snRNP