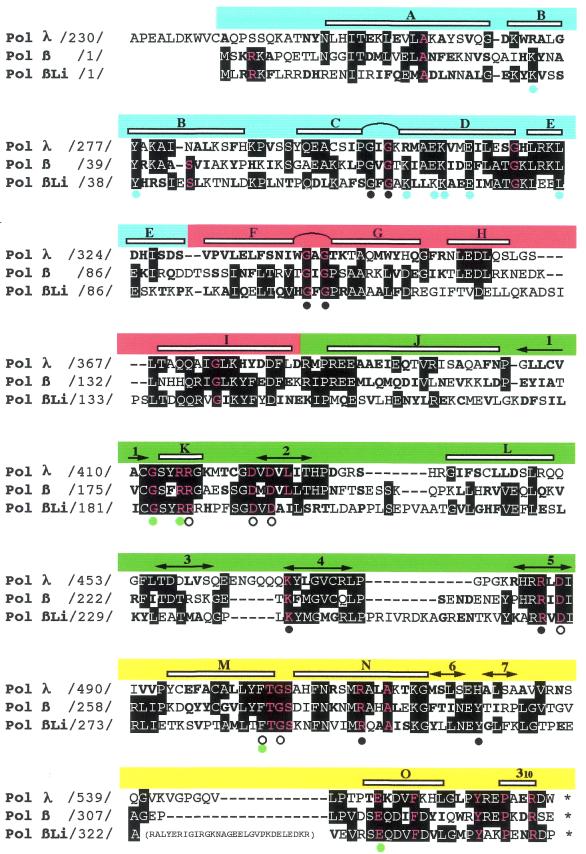
Figure 1.
Li Pol β, a Leishmania nuclear polymerase homologous to Pol β. Multiple amino acid alignment of mouse Pol λ, rat Pol β and Li Pol β. Numbers between slashes indicate the amino acid position relative to the N-terminus of each DNA polymerase. According to rat Pol β structural data (15,29), the alignment can be divided in four subdomains: ‘8 kDa’ (residues 1–84; blue segment), ‘fingers’ (residues 85–153; red segment), ‘palm’ (residues 154–276; green segment) and ‘thumb’ (residues 277–376; yellow segment). α-helices (lettered) and β-strands (numbered) are indicated at the top of the alignment. Two helix–hairpin–helix motifs are between α-helices C and D (‘8 kDa’ subdomain), and between α-helices F and G (‘fingers’ subdomain). Invariant residues among any pair of DNA polymerases are indicated with white letters (over a black background). Conservative substitutions (bold typed) were considered as follows: K, H and R; D, E, Q and N; W, F, Y, I, L, V, M and A; G, S, T, C and P. The 27 residues that are invariant among DNA polymerase X family members (8) are indicated with red letters. Colored dots at the bottom of the alignment indicate the Pol β residues (15) shown to act as DNA ligands (Gly64, Gly66, Gly105, Gly109, Lys234, Arg254, Arg283, Tyr296; black), dNTP and metal ligands (Arg183; Asp190, Asp192, Asp256, Phe272 and Gly274; white), or involved in interactions between the ‘palm’ and ‘thumb’ subdomains (Gly179/Phe272; Arg182/Glu316; green). Blue dots indicate residues involved in the dRPase activity (Lys35, Tyr39, Lys68, Glu71, Lys72, Glu75 and Lys84) present at the ‘8 kDa’ domain of Pol β (66–68). A 29 amino acid insertion in Li Pol β, located between β-strand 7 and helix O, is shown in parentheses.