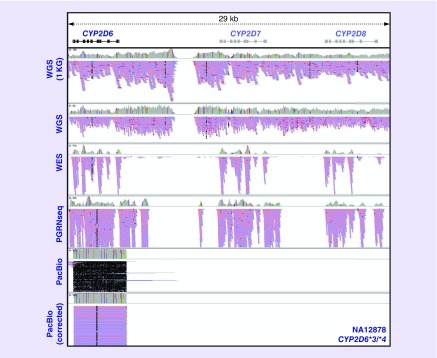
Figure 4. . Paired-end short-read sequencing (Illumina) and long-read sequencing (Pacific Biosciences) of the CYP2D6 gene region visualized with the Integrative Genomics Viewer.
Results for NA12878 (CYP2D6*3/*4) are displayed from top to bottom panels for WGS from the 1000 Genomes Project, in-house WGS, WES, targeted capture with the PGRNseq platform, targeted PacBio CYP2D6 sequencing, and ALEC-corrected targeted PacBio CYP2D6 sequencing. Of note, discrepant and skewed allele frequencies in several loci from the WGS data indicate potential read misalignment errors. Moreover, the common CYP2D6 capture strategies (e.g., WES, PGRNseq) coupled with short-read Illumina sequencing result in significant read assignment to the CYP2D7 and CYP2D8 pseudogenes. These reads indicate a lack of specificity for CYP2D6 by these target enrichment approaches and/or informatic errors related to read misalignment. Targeted PacBio sequencing results in CYP2D6-specific sequencing and no misalignment to CYP2D7 or CYP2D8, but random errors throughout the sequencing reads are characteristic to this technology. These random errors can be minimized by circular consensus sequencing read analysis; however, further correction prior to variant calling can also be accomplished by available informatics tools (e.g., Amplicon Long-read Error Correction [ALEC]).
1KG: 1000 Genomes Project; PacBio: Pacific Biosciences; WES: Whole-exome sequencing; WGS: Whole-genome sequencing.