Fig. 4.
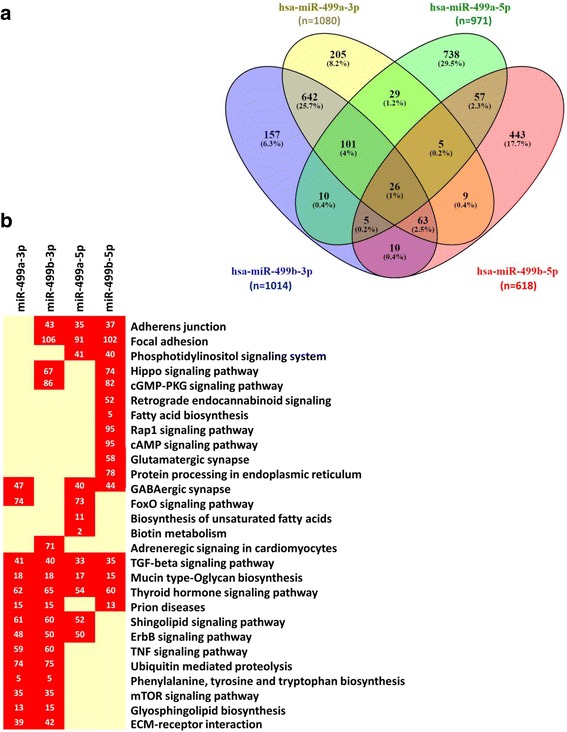
Enrichment pathway analysis of MIR-499 gene cluster targets. (a) Venn diagram showing the intersection of gene targets for each microRNA. Gene targets were retrieved from microT-CDS Diana tools (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=microT_CDS/index) with threshold set at 0.7 and significance <0.05. Venn diagram was drawn using Venny v2.0 (http://bioinfogp.cnb.csic.es/tools/venny/). The figure shows that hundreds of gene targets are complementary to miR-499a and miR-499b. Some of these genes are commonly targeted by both genes, showing some similarities in their function. However, other gene targets that are distinct for each mature form, indicate the presence of unique function for each of them. (b) Heat map showing top significant KEGG pathways involved by miRNA targets. Result intersection and filtration after FDR correction was applied. Fisher’s Exact test (hypergeometeric distribution) was used for statistical analysis. Heat map represent some of the significant KEGG pathways related to the target gene set of each mature miRNA. The number in the heat map represents the number of gene targets in each pathway affected by the corresponding miRNA. The four mature forms of MIR-499 gene family is involved in TGF-beta signaling pathway and mucin biosynthesis. Additionally, miR-499a-5p along with miR-499b are enrolled in adherence junction and focal adhesion
