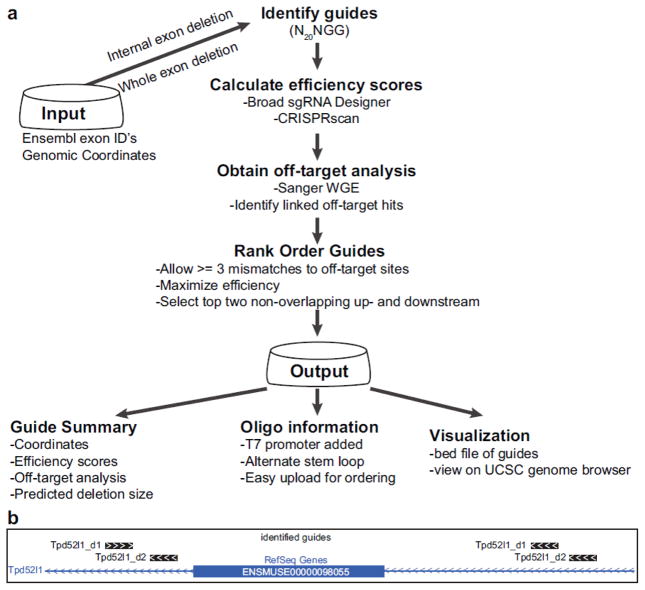
Figure 2.
CRISPRtools workflow for designing null alleles. (a) Critical exon identifiers or genomic coordinates are used as input and rank ordered guides are provided as output with a series of summary files containing guide positions, scores, expected deletion sizes, oligo information, and a bed file for visualization on genome browser. (b) Stylized representation of UCSC genome browser view illustrating exon deletion strategy using four guides.