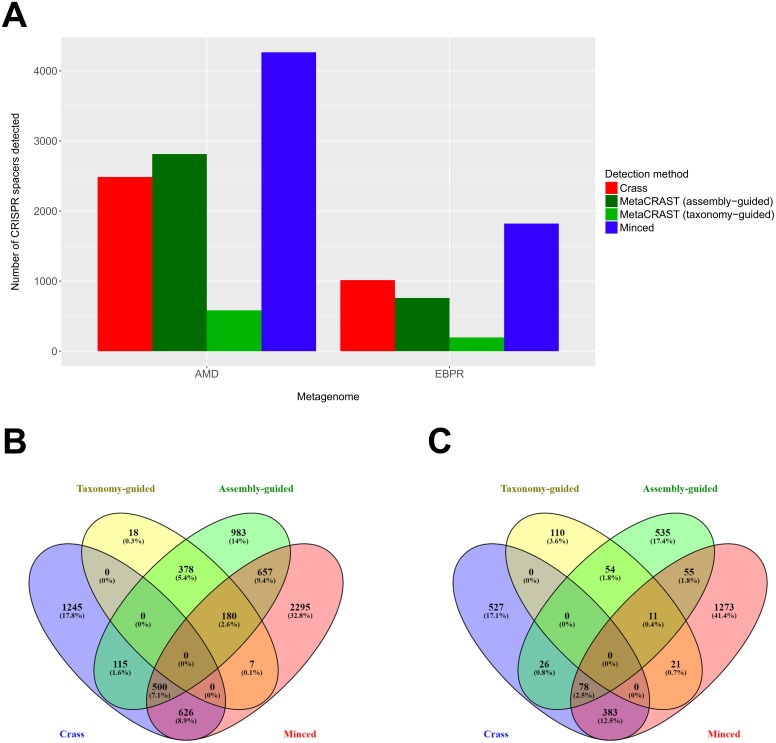
Figure 5. Evaluation of MetaCRAST, Crass, and MinCED on real AMD and EBPR metagenomes.
(A) Total number of CRISPR spacers detected in real AMD and EBPR metagenomes using four different detection methods—Crass (de novo), MetaCRAST (using assembly-guided queries), MetaCRAST (using taxonomy-guided queries), and MinCED (de novo). Taxonomy-guided and assembly-guided queries are provided as Tables S3–S4 and S6–S7. (B) Comparison of spacers detected in the real AMD metagenome using Crass (de novo), MetaCRAST (using taxonomy-guided queries), MetaCRAST (using assembly-guided queries), and MinCED (de novo). Comparison was performed using Venny 2.1 (http://bioinfogp.cnb.csic.es/tools/venny/). (C) Comparison of spacers detected in the real EBPR metagenome using the same methods as in (B) Comparison was performed using Venny 2.1.