Abstract
Enterobacter sp. DMKU-RP206 was isolated from rice leaves in Thailand and identified by the 16S rRNA gene and multilocus sequence (gyrB, rpoB, atpD, and infB genes) analysis. The bacterium was assessed on plant growth-promoting traits including indole-3-acetic acid (IAA) production. Phosphate solubilization, ammonia production, and antagonism to fungal plant pathogens, as well as siderophore production, were shown by this bacterium. However, only IAA production was focused on. The production of IAA by Enterobacter sp. DMKU-RP206 was optimized by statistical methods. A Box–Behnken design was used for the investigation of interactions among the basic influencing factors and for the optimization of IAA production. The results showed that l-tryptophan had a significant importance in terms of IAA production. Enterobacter sp. DMKU-RP206 produced a higher amount of IAA than previously reported for the genus Enterobacter. 0.85% of lactose as a carbon source, 1.3% of yeast extract as a nitrogen source, 1.1% of l-tryptophan as a precursor, 0.4% of NaCl, an initial pH of 5.8, an incubation temperature at 30 °C, and a shaking speed of 200 rpm were found to be the optimum conditions for IAA production. In addition, IAA production was performed to scale up IAA production, and the highest amount, 5561.7 mg l−1, was obtained. This study reported a 13.4-fold improvement in IAA production by Enterobacter sp. DMKU-RP206.
Keywords: Enterobacter sp., Indole-3-acetic acid, Multilocus sequence analysis, Optimization, Phyllosphere bacteria, Plant growth-promoting traits
Introduction
Indole-3-acetic acid (IAA) is a plant hormone that is classified as being part of the auxin family. The roles of IAA have been reported in plants and microorganisms. In plants, IAA stimulates root initiation, lateral root formation, cell division, cell elongation, cell differentiation, and light and gravitational responses, including the regulation of falling leaves and fruit ripening. It also offers increased protection to plants in terms of external stress (Teale et al. 2006). Various roles of IAA have been reported in microorganisms such as the stimulation of spore germination and mycelial elongation in Streptomyces (Matsukawa et al. 2007), induced filamentation and substrate adhesion in Saccharomyces cerevisiae (Prusty et al. 2004), and stimulated filamentation in Candida albicans (Rao et al. 2010). Literature reviews indicate that many plant-associated microorganisms such as bacteria (Tsavkelova et al. 2007a) and yeasts (Nutaratat et al. 2014) possess an IAA-producing ability. Apart from plant hormones, plant-associated microorganisms can promote plant growth by the inhibition of plant ethylene synthesis through 1-aminocyclopropane-1-carboxylate (ACC) deaminase activity. It can also promote the improvement of nutrient uptake through inorganic phosphate and zinc oxide solubilization, and through ammonia and siderophore production (Nadeem et al. 2016; Scagliola et al. 2016; Shaikh and Saraf 2017). In addition, they are able to decrease or prevent the deleterious effects of pathogenic microorganisms through siderophore and hydrogen cyanide production, including cell wall degrading enzyme activities (de Souza et al. 2015; Fu et al. 2016).
IAA-producing bacteria have been reported in several studies. These belong to the genera Agrobacterium, Burkholderia, Bacillus, Erwinia, Flavobacterium, Pantoea, Pseudomonas (Tsavkelova et al. 2007a), Microbacterium, Mycobacterium, Rhizobium, and Sphingomonas (Tsavkelova et al. 2007b), including Enterobacter (Shoebitz et al. 2009; Jasim et al. 2014; Ghosh et al. 2015). The genus Enterobacter is found in various environments in association with plants such as winter wheat phyllosphere (Kämpfer et al. 2005), winter ryegrass rhizosphere (Shoebitz et al. 2009), corn rhizosphere (Kämpfer et al. 2015), and tobacco stem (Duan et al. 2015). There have been many reports on Enterobacter that support plant growth. For example, E. ludwigii BNM 0357 improved the development of the winter ryegrass (Lolium perenne) root system (Shoebitz et al. 2009). The data of Shahid et al. (2012) showed that Enterobacter sp. Fs-11 is an efficient phosphate solubilizing bacteria and it has high potential in terms of being used as a bio-inoculant for sunflowers under phosphorus-deficient conditions. It is indicated that Enterobacter is one of the bacterial resources suitable for agricultural use. Even though IAA production has been reported from various Enterobacter spp., the quantitative analysis of IAA and its maximum possible yield have not been well studied. In addition, there have only been a few reports on the optimization of IAA-producing Enterobacter. Therefore, we have employed statistical methods to investigate and report on the optimization of IAA production in Enterobacter sp. DMKU-RP206. The validation of IAA production on a large scale is also reported.
Materials and methods
Microorganism, identification, and cultivation
The bacterium strain DMKU-RP206, efficient for IAA production, was isolated from rice leaves in Thailand using the plating of leaf washings technique, as described by Inácio et al. (2002). Tryptone soy agar (TSA) (17 g l−1 tryptone, 3 g l−1 soy peptone, 5 g l−1 sodium chloride, 2.5 g l−1 dipotassium hydrogen phosphate, 2.5 g l−1 dextrose, and 15 g l−1 agar) was used to maintain the bacterial strain.
This bacterium was identified by analysis of similarities in the 16S rRNA gene sequence by using universal primers 27F and 1525R (Lane 1991). The 16S rRNA gene sequence of the strain DMKU-RP206 was compared with that of the type strain from the EzBioCloud database (http://www.ezbiocloud.net/) (Yoon et al. 2017). In addition, multilocus sequence analysis (MLSA) based on gyrB, rpoB, atpD, and infB genes was performed. The gyrB gene was amplified using universal primers as described by Yamamoto and Harayama (1995). The amplification and sequencing of DMKU-RP206 rpoB, atpD, and infB genes were performed as described by Brady et al. (2008). The obtained sequences were compared with closely related strains from the GenBank database (http://www.ncbi.nlm.nih.gov). Phylogenetic trees based on 16S rRNA gene sequences were constructed from the evolutionary distance data with a Kimura 2-parameter model (Kimura 1980), using the neighbor-joining method (Saitou and Nei 1987) with the program MEGA (molecular evolutionary genetics analysis) version 6 (Tamura et al. 2013) software. Bootstrap confidence values were obtained using 1000 resamplings (Felsenstein 1985). The sequence of strain DMKU-RP206 was aligned with their closest types of strains of species using MUSCLE (multiple sequence comparison by log-expectation) (Edgar 2004).
Characterization of plant growth promoting traits of the bacterium strain DMKU-RP206
Direct mechanisms
Phosphate and zinc oxide solubilization, ammonia and polyamine production, and ACC deaminase activity were described as direct mechanisms by Zaidi et al. (2006), Rokhbakhsh-Zamin et al. (2011), Cappuccino and Sherman (1992), Cloete et al. (2009), and Dell’Amico et al. (2005), respectively. The bacterium strain DMKU-RP206 grown in a tryptone soy medium at 30 ± 2 °C for 15 h was used as an inoculum for the characterization of plant growth-promoting traits. The inoculum was spotted onto Pikovskaya’s agar (for phosphate solubilization), zinc oxide agar (for zinc oxide solubilization), and Long Ashton decarboxylase (LAD) agar (for polyamine production), and was transferred to tryptic soy broth (TSB) supplemented with 0.1% l-tryptophan (for IAA production), peptone water (for ammonia production), and a Dworkin–Foster minimal salts medium supplemented with 3 mM ACC (for ACC deaminase activity). All experiments were incubated at 30 ± 2 °C for 5–7 days and performed with three replicates.
Indirect mechanisms
Indirect characters of plant growth promotion were studied. Siderophore production was determined (Schwyn and Neilands 1987) as well as hydrogen cyanide production (Bakker and Schipper 1987). Cell wall degrading enzyme activities—i.e. cellulose (Paudel and Qin 2015), chitinase (Sato et al. 2009), and protease (Cattelan et al. 1999) were also investigated using Chrome Azurol S agar, TSA amended glycine, carboxymethyl cellulose (CMC) agar, a chitin medium, and a skimmed milk medium, respectively. In addition, antagonistic activity against phytopathogenic fungi was evaluated by in vitro tests using a dual-culture technique (Vaikuntapu et al. 2014). Rice fungal pathogens such as Curvularia lunata, Fusarium moniliforme, and Rhizoctonia solani were used in this study. Percent inhibition was determined by percent inhibition = [(R – r)/R] × 100, where R is the radius of the fungal colony away from the bacterial colony and r is the length of the fungal colony opposite the bacterial colony. All experiments were incubated at 30 ± 2 °C for 5–7 days and three experimental replicates were performed.
Optimization of IAA production
Effect of incubation time on IAA production
Bacterial inoculum was prepared in 250 ml Erlenmeyer flasks containing 50 ml of TSB (TSA without agar) and incubated on a rotary shaker at 150 rpm at 30 ± 2 °C for 16–18 h. The culture was transferred into 50 ml of TSB supplemented with 0.1% (w/v) l-tryptophan. The initial OD600 of the working culture was adjusted to 0.10–0.15 prior to incubation on an orbital shaker at 150 rpm at 37 °C for 10 days. Samples were taken daily, and culture supernatants were collected by centrifugation for 5 min at 5000×g. The effect of the incubation time on IAA production was performed in triplicate. IAA in the culture supernatants was analyzed by high-performance liquid chromatography (HPLC) (Agilent Technologies, USA) equipped with a Cosmosil SC18-MS-II column (Nacalai Tesque, Japan) and a UV-detector at 280 nm. A mixture of 40% of solution A (methanol:acetic acid:water; 10:0.3:89.7 v/v/v) and 60% of solution B (methanol:acetic acid:water; 90:0.3:9.7 v/v/v) was used as the mobile phase, with a flow rate of 0.5 ml min−1, as described by Nutaratat et al. (2014).
Preliminary screening of optimal carbon and nitrogen sources on IAA production
Bacterial inoculum was prepared in 125 ml Erlenmeyer flasks containing 25 ml of TSB and incubated on a rotary shaker at 150 rpm at 30 ± 2 °C for 16–18 h. The culture was transferred into a 5-ml medium broth containing a 1% carbon source and a 0.1% nitrogen source amended with 0.1% (w/v) l-tryptophan. The effects of various carbon and nitrogen sources on IAA production were assessed by altering the composition of the medium accordingly. Various carbon sources (arabinose, dextrose, fructose, galactose, glycerol, lactose, maltose, mannitol, mannose, my-inosital, raffinose, sorbitol, sorbose, starch, sucrose, xylitol, and xylose) and various nitrogen sources (NH4Cl, (NH4)2SO4, corn steep liquor, malt extract, monosodium glutamate, peptone, KNO3, skimmed milk, soy isolate, NaNO2, tryptone, urea, and yeast extract) were investigated. The initial OD600 was adjusted to 0.1–0.15 prior to incubation on a reciprocal shaker at 150 strokes min−1 at 30 ± 2 °C for optimal incubation time. The culture supernatant was collected by centrifugation for 5 min at 5000×g. The IAA in the culture supernatants was analyzed using the colorimetric method. The assay was carried out by mixing 1 ml of culture supernatant with 1 ml of Salkowski reagent containing 12 g l−1 FeCl3 and 7.9 mol l−1 H2SO4 (Glickmann and Dessaux 1994), followed by 30 min incubation in the dark to develop a pink-red color. The color intensity was determined as A530 using a spectrophotometer (Thermo Spectronic, USA). The IAA concentration of the sample was compared with a linear calibration curve obtained from an authentic IAA standard. All experiments were done in triplicate.
Optimization of IAA production using statistical methods
In this study, we focused on the basic influencing factors in terms of IAA production, i.e. carbon source, nitrogen source, IAA precursor (l-tryptophan), NaCl, initial pH, temperature, and shaking speed. The Box–Behnken design was used to investigate the interaction among the influencing factors and in order to optimize IAA production. A total of seven factors are shown in Table 1. A total of 62 treatments were carried out in 250 ml Erlenmeyer flasks containing 50 ml of the medium in triplicate. The IAA in the culture supernatants was analyzed by HPLC.
Table 1.
Box–Behnken experimental design for IAA production by Enterobacter sp. DMKU-RP206
| Treatments | Factorsa | IAA (mg l−1) | ||||||
|---|---|---|---|---|---|---|---|---|
| X 1 | X 2 | X 3 | X 4 | X 5 | X 6 | X 7 | ||
| 1 | 0.1 | 1.0 | 0.9 | 2.0 | 7 | 35 | 200 | 1223.9 ± 44.9 |
| 2 | 0.8 | 0.5 | 0.9 | 2.0 | 8 | 30 | 100 | 1578.9 ± 19.4 |
| 3 | 1.5 | 1.5 | 0.9 | 1.0 | 7 | 30 | 150 | 2227.1 ± 46.1 |
| 4 | 0.8 | 1.5 | 0.9 | 2.0 | 8 | 30 | 200 | 1613.0 ± 31.2 |
| 5 | 1.5 | 1.0 | 1.3 | 2.0 | 8 | 30 | 150 | 1145.9 ± 22.1 |
| 6 | 0.8 | 1.0 | 0.9 | 1.0 | 8 | 25 | 150 | 1229.2 ± 57.1 |
| 7 | 0.1 | 1.5 | 0.9 | 1.0 | 7 | 30 | 150 | 1092.4 ± 36.9 |
| 8 | 0.8 | 1.0 | 0.9 | 3.0 | 6 | 25 | 150 | 666.1 ± 3.2 |
| 9 | 0.8 | 0.5 | 0.9 | 2.0 | 6 | 30 | 100 | 1729.9 ± 26.1 |
| 10 | 0.1 | 0.5 | 0.9 | 1.0 | 7 | 30 | 150 | 931.1 ± 29.5 |
| 11 | 0.1 | 1.5 | 0.9 | 3.0 | 7 | 30 | 150 | 838.8 ± 13.7 |
| 12 | 1.5 | 1.0 | 0.5 | 2.0 | 6 | 30 | 150 | 1380.1 ± 27.1 |
| 13 | 0.8 | 1.0 | 0.9 | 3.0 | 6 | 35 | 150 | 749.7 ± 43.1 |
| 14 | 0.8 | 1.0 | 0.5 | 1.0 | 7 | 30 | 100 | 1413.0 ± 5.9 |
| 15 | 0.1 | 1.0 | 1.3 | 2.0 | 8 | 30 | 150 | 238.9 ± 24.2 |
| 16 | 1.5 | 1.0 | 1.3 | 2.0 | 6 | 30 | 150 | 1817.6 ± 101.7 |
| 17 | 1.5 | 1.5 | 0.9 | 3.0 | 7 | 30 | 150 | 2029.7 ± 39.6 |
| 18 | 0.8 | 1.0 | 0.5 | 1.0 | 7 | 30 | 200 | 2436.7 ± 35.4 |
| 19 | 0.8 | 0.5 | 0.5 | 2.0 | 7 | 25 | 150 | 482.0 ± 4.8 |
| 20 | 0.8 | 1.0 | 0.9 | 2.0 | 7 | 30 | 150 | 2969.0 ± 17.9 |
| 21 | 0.1 | 1.0 | 0.5 | 2.0 | 8 | 30 | 150 | 689.6 ± 14.6 |
| 22 | 0.1 | 1.0 | 0.9 | 2.0 | 7 | 25 | 100 | 660.6 ± 21.4 |
| 23 | 0.8 | 1.0 | 0.9 | 2.0 | 7 | 30 | 150 | 3059.4 ± 72.2 |
| 24 | 0.8 | 0.5 | 0.9 | 2.0 | 6 | 30 | 200 | 1520.5 ± 16.0 |
| 25 | 0.1 | 1.0 | 0.9 | 2.0 | 7 | 35 | 100 | 1060.4 ± 14.6 |
| 26 | 0.8 | 1.5 | 0.5 | 2.0 | 7 | 25 | 150 | 1352.9 ± 20.5 |
| 27 | 0.8 | 1.0 | 0.5 | 3.0 | 7 | 30 | 100 | 1407.7 ± 8.8 |
| 28 | 0.8 | 1.0 | 0.9 | 2.0 | 7 | 30 | 150 | 2875.2 ± 26.8 |
| 29 | 0.8 | 1.0 | 0.9 | 2.0 | 7 | 30 | 150 | 2857.4 ± 17.7 |
| 30 | 1.5 | 1.0 | 0.5 | 2.0 | 8 | 30 | 150 | 1396.6 ± 26.6 |
| 31 | 0.8 | 0.5 | 1.3 | 2.0 | 7 | 35 | 150 | 646.3 ± 4.0 |
| 32 | 0.8 | 1.0 | 0.9 | 3.0 | 8 | 35 | 150 | 1181.2 ± 20.2 |
| 33 | 0.8 | 1.5 | 0.9 | 2.0 | 8 | 30 | 100 | 1761.5 ± 69.6 |
| 34 | 0.8 | 0.5 | 0.5 | 2.0 | 7 | 35 | 150 | 1951.6 ± 80.0 |
| 35 | 0.8 | 1.0 | 0.9 | 2.0 | 7 | 30 | 150 | 2826.1 ± 48.6 |
| 36 | 0.1 | 1.0 | 1.3 | 2.0 | 6 | 30 | 150 | 1350.6 ± 13.9 |
| 37 | 1.5 | 1.0 | 0.9 | 2.0 | 7 | 35 | 100 | 291.2 ± 8.2 |
| 38 | 0.8 | 1.0 | 1.3 | 3.0 | 7 | 30 | 200 | 65.1 ± 5.2 |
| 39 | 0.8 | 1.0 | 0.9 | 2.0 | 7 | 30 | 150 | 2975.5 ± 64.5 |
| 40 | 0.1 | 1.0 | 0.5 | 2.0 | 6 | 30 | 150 | 853.1 ± 30.1 |
| 41 | 0.8 | 1.0 | 1.3 | 3.0 | 7 | 30 | 100 | 219.6 ± 3.8 |
| 42 | 0.8 | 1.5 | 0.9 | 2.0 | 6 | 30 | 200 | 2867.3 ± 30.3 |
| 43 | 0.8 | 1.0 | 0.9 | 1.0 | 6 | 35 | 150 | 2175.7 ± 31.2 |
| 44 | 1.5 | 1.0 | 0.9 | 2.0 | 7 | 25 | 100 | 1429.5 ± 61.0 |
| 45 | 0.8 | 1.5 | 0.9 | 2.0 | 6 | 30 | 100 | 1936.6 ± 33.7 |
| 46 | 0.8 | 1.0 | 0.5 | 3.0 | 7 | 30 | 200 | 1237.7 ± 33.6 |
| 47 | 0.1 | 1.0 | 0.9 | 2.0 | 7 | 25 | 200 | 1025.2 ± 23.2 |
| 48 | 1.5 | 1.0 | 0.9 | 2.0 | 7 | 35 | 200 | 316.5 ± 21.6 |
| 49 | 1.5 | 0.5 | 0.9 | 3.0 | 7 | 30 | 150 | 1100.0 ± 53.0 |
| 50 | 0.1 | 0.5 | 0.9 | 3.0 | 7 | 30 | 150 | 760.2 ± 7.8 |
| 51 | 0.8 | 1.0 | 0.9 | 3.0 | 8 | 25 | 150 | 953.5 ± 4.3 |
| 52 | 0.8 | 1.5 | 1.3 | 2.0 | 7 | 25 | 150 | 996.3 ± 34.9 |
| 53 | 0.8 | 1.5 | 0.5 | 2.0 | 7 | 35 | 150 | 1401.8 ± 59.4 |
| 54 | 0.8 | 1.0 | 0.9 | 1.0 | 8 | 35 | 150 | 2097.2 ± 103.9 |
| 55 | 1.5 | 0.5 | 0.9 | 1.0 | 7 | 30 | 150 | 586.5 ± 27.0 |
| 56 | 0.8 | 0.5 | 0.9 | 2.0 | 8 | 30 | 200 | 1373.3 ± 11.2 |
| 57 | 1.5 | 1.0 | 0.9 | 2.0 | 7 | 25 | 200 | 1592.3 ± 63.9 |
| 58 | 0.8 | 1.0 | 1.3 | 1.0 | 7 | 30 | 200 | 3058.1 ± 3.8 |
| 59 | 0.8 | 0.5 | 1.3 | 2.0 | 7 | 25 | 150 | 799.4 ± 28.4 |
| 60 | 0.8 | 1.0 | 0.9 | 1.0 | 6 | 25 | 150 | 2237.9 ± 12.4 |
| 61 | 0.8 | 1.0 | 1.3 | 1.0 | 7 | 30 | 100 | 3158.8 ± 72.2 |
| 62 | 0.8 | 1.5 | 1.3 | 2.0 | 7 | 35 | 150 | 550.3 ± 33.3 |
a X 1 lactose (%), X 2 yeast extract (%), X 3 l-tryptophan (%), X 4 NaCl (%), X 5 initial pH, X 6 temperature (°C), X 7 shaking speed (rpm)
The data with regard to the IAA concentration were subjected to multiple linear regression analysis using Statistica Software version 10.0 (StatSoft Inc, USA). The model coefficients and their standard errors were calculated. The contour plots were drawn and the optimum concentrations of the factors were determined for maximum IAA production using Statistica Software version 10.0. Finally, the model and the values obtained were validated in triplicate.
IAA production in 2 l stirred tank fermenter
50 ml (3.5%) of bacterial inoculum (OD600 ≈ 2.9–3.0) was transferred to 1.45 l of the optimized production medium. The bioreactor (B. Braun Biotech International, Germany) was operated under conditions by which the agitation speed was fixed at 200 rpm and the aeration rate was varied at 1, 2, and 3 l min−1 (equivalent to 0.67, 1.33 and 2 vvm, respectively). The medium pH was uncontrolled throughout the course of the fermentation and was similar in all cultures performed in the shake flasks. The culture pH was measured by pH electrodes (Mettler Toledo, Switzerland). Samples were taken daily for 7 days. The supernatant was collected by centrifugation and analyzed for IAA by HPLC. All experiments were done in duplicate.
Results
Identification of the phyllosphere bacterium strain DMKU-RP206
Comparative analysis of the16S rRNA gene sequence of the strain DMKU-RP206 (1507 bp; GenBank accession number MF125281), with the type of strain of the genus Enterobacter, revealed that strain DMKU-RP206 showed the highest level of 16S rRNA gene similarity with the type of strain of Enterobacter tabaci YIM Hb-3T (99.66%) followed by E. muelleri JM-458T (99.43%) and E. mori R18-2T (99.18%). The phylogenetic tree based on 16S rRNA gene sequences (Fig. 1a) indicated that strain DMKU-RP206 clustered within the genus Enterobacter with its closest species, i.e. E. tabaci YIM Hb-3T and E. muelleri JM-458T, while, MLSA based on gyrB (742 bp; MF197444), rpoB (637 bp; MF197445), atpD (642 bp; MF197446), and infB (615 bp; MF197447) gene sequences (Fig. 1b) showed that strain DMKU-RP206 clustered with its closest species, i.e. E. muelleri JM-458T, E. cloacae subsp. cloacae LMG 2783T, and E. cloacae subsp. dissolvens LMG 2683T. However, both of phylogenetic trees suggested that strain DMKU-RP206 formed a distinct branch within the genus Enterobacter. Thus, in this study, the bacterium strain DMKU-RP206 was identified as Enterobacter.
Fig. 1.
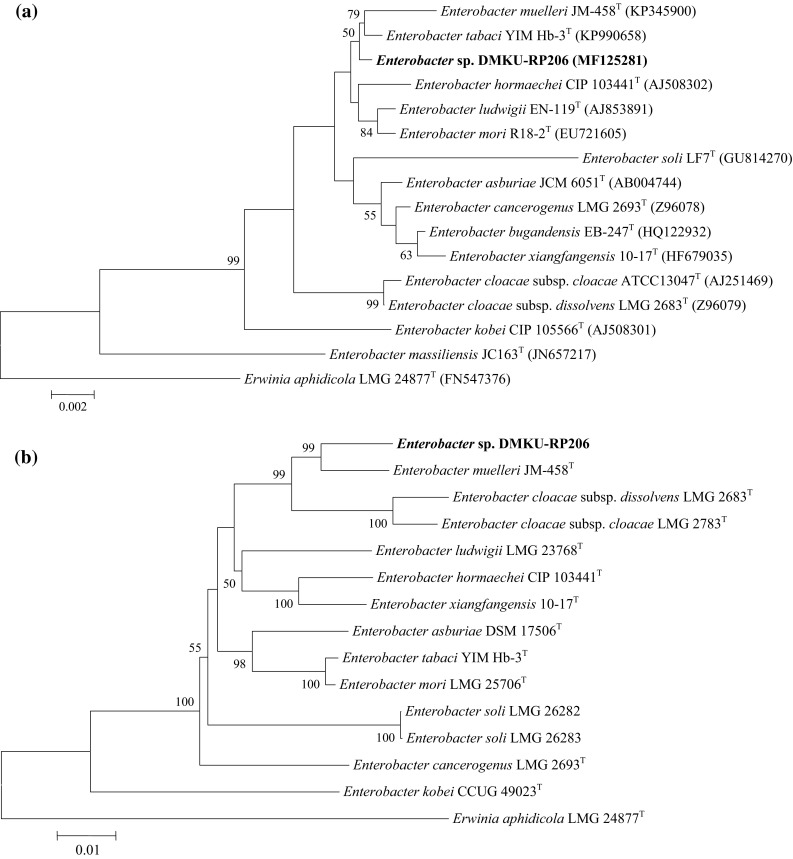
The phylogenetic relationships between strain DMKU-RP206 and the type strains of genus Enterobacter base on 16S rRNA gene sequences (a) and concatenated partial gyrB, rpoB, atpD, and infB gene sequences (b). The sequence of Erwinia aphidicola LMG 24877T was used as an outgroup. Bootstrap values below 50% are not indicated
Plant growth promoting traits of Enterobacter sp. DMKU-RP206
Enterobacter sp. DMKU-RP206 showed the capability of phosphate solubilization and IAA, ammonia, and siderophore production, including fungal antagonistic activity. However, other plant growth promoting traits—i.e. zinc oxide solubilization, ACC deaminase activity, polyamine and hydrogen cyanide production, and cell wall degrading enzyme activities (cellulose, chitinase, and protease)—were not detected in this bacterium. Among the positive activities of the plant growth-promoting traits, ammonia and siderophore production together with phosphate solubilization showed slight activities. Noticeably, antagonistic activity exhibited a satisfactory result (Fig. 2). This bacterium showed the highest inhibition of the growth of fungal rice pathogens C. lunata (56.4%), followed by R. solani (53.8%) and F. moniliforme (19.0%). Although the bacterium Enterobacter sp. DMKU-RP206 exhibited some plant growth-promoting characteristics, it was shown to be outstanding in IAA production. The following study was thus focused solely on its IAA production.
Fig. 2.
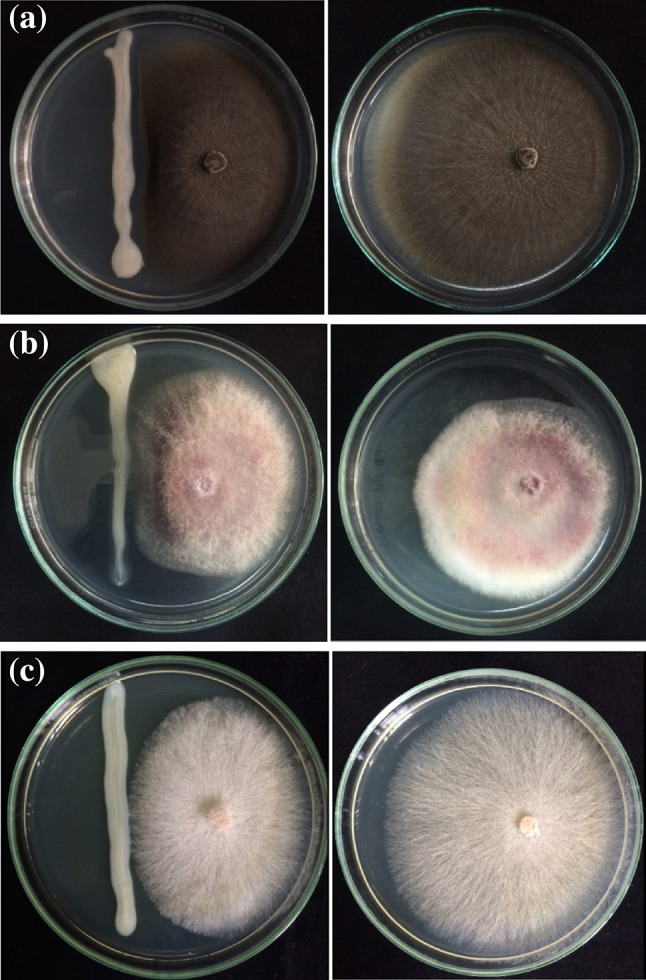
Antagonism of Enterobacter sp. DMKU-RP206 against Curvularia lunata (a), Fusarium moniliforme (b), and Rhizoctonia solani (c), which causes disease in rice plants
Optimization of IAA production by Enterobacter sp. DMKU-RP206
Effect of incubation time
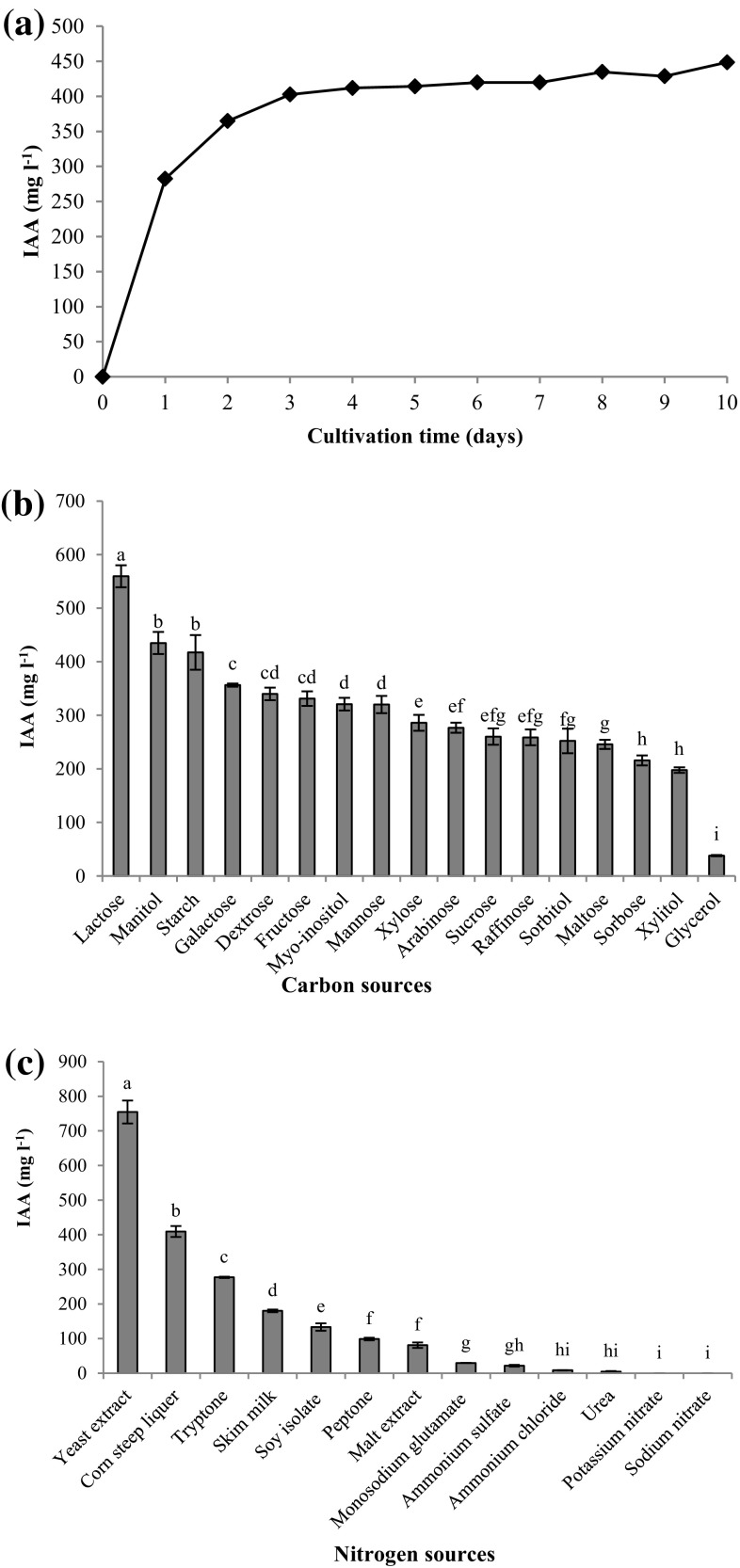
The Enterobacter sp. DMKU-RP206 started to produce IAA at a rate of 282.4 mg l−1 after 24 h of incubation. The maximum amount of IAA (448.5 mg l−1) was observed at 10 days of incubation (Fig. 3a). However, Fig. 3a also indicates that slightly lower amounts of IAA were detected after 5 days of incubation. When production cost is considered, we decided to choose an incubation time of 5 days when 414.2 mg IAA l−1 was obtained in order to reduce the production time.
Fig. 3.
Effect of incubation time (a), carbon sources (b), and nitrogen sources (c) on IAA production by Enterobacter sp. DMKU-RP206. Bars with different letters are significantly different from each other (p < 0.05)
Preliminary screening of optimal carbon and nitrogen sources
For optimal carbon source screening, the replacement of dextrose in a basal medium, TSB, with 17 varied carbon sources, revealed that Enterobacter sp. DMKU-RP206 utilized various carbon sources for IAA production (Fig. 3b). Among the 17 carbon sources tested, lactose was found to be the best choice for IAA production, followed by mannitol and starch. Enterobacter sp. DMKU-RP206 produced the highest amount of IAA (559.6 mg l−1) when cultivated in a medium containing lactose. Regarding the nitrogen sources, the replacement of tryptone and soy peptone in TSB with 13 different nitrogen sources showed that the highest amount of IAA produced (754.4 mg l−1) was observed when yeast extract was used (Fig. 3c).
Optimization of IAA production using statistical methods
The Box–Behnken design was used to determine the optimal levels of influencing factors with regard to IAA production. The average IAA production from 65.1 ± 5.2 to 3158.8 ± 72.2 mg l−1 was obtained (Table 1). The experimental data were subjected to analysis of variance (ANOVA) and multiple linear regressions as shown in Tables 2 and 3, respectively. The p value for the model (p < 0.0001) and the coefficient of determination (R 2 = 0.85) suggest that the variability in the response can be accounted for by the model. The second-order polynomial model for IAA production is as follows:
where Y is indole-3-acetic acid (mg l−1), X 1 is lactose, X 2 is yeast extract, X 3 is l-tryptophan, X 4 is NaCl, X 5 is the initial pH, X 6 is temperature, and X7 is the shaking speed.
Table 2.
Analysis of variance (ANOVA) in the regression model for optimization of IAA production by Enterobacter sp. DMKU-RP206
| SOV | Sum of squares | Degree of freedom | Mean square | F-value | p-value |
|---|---|---|---|---|---|
| Model | 105355341 | 35 | 3010153 | 24.48 | <0.0001 |
| Residual | 18444964 | 150 | 122966 | ||
| Total | 123800305 | 185 |
R 2 = 0.85; R 2 adjust = 0.82
Table 3.
Coefficient and p-value of regression analysis for IAA production by Enterobacter sp. DMKU-RP206
| Model | Coefficient | p-value |
|---|---|---|
| Constant | −63624.5 | <0.0001a |
| X 1 (lactose) | 4582.3 | <0.0001a |
| X 2 (yeast extract) | 7826.7 | <0.0001a |
| X 3 (l-tryptophan) | 18,352.4 | <0.0001a |
| X 4 (sodium chloride) | 1715.7 | 0.0264a |
| X 5 (pH) | 4318.8 | <0.0001a |
| X 6 (temperature) | 2079.0 | <0.0001a |
| X 7 (rpm) | 69.7 | <0.0001a |
| X 1 X 2 | 832.3 | 0.0001a |
| X 1 X 3 | 61.9 | 0.8091 |
| X 1 X 4 | 132.3 | 0.1978 |
| X 1 X 5 | 111.0 | 0.2795 |
| X 1 X 6 | −107.6 | <0.0001a |
| X 1 X 7 | −1.2 | 0.5536 |
| X 2 X 3 | −137.7 | 0.7010 |
| X 2 X 4 | −198.4 | 0.1678 |
| X 2 X 5 | −282.8 | 0.0500a |
| X 2 X 6 | −85.7 | 0.0032a |
| X 2 X 7 | 6.0 | 0.0382a |
| X 3 X 4 | −1477.5 | <0.0001a |
| X 3 X 5 | −511.0 | 0.0049a |
| X 3 X 6 | −132.4 | 0.0003a |
| X 3 X 7 | −6.9 | 0.0547 |
| X 4 X 5 | 225.8 | 0.0019a |
| X 4 X 6 | −12.4 | 0.3892 |
| X 4 X 7 | −3.1 | 0.0309a |
| X 5 X 6 | 26.9 | 0.0626 |
| X 5 X 7 | −2.7 | 0.0623 |
| X 6 X 7 | −0.2 | 0.5553 |
| X 21 | −1768.1 | <0.0001a |
| X 22 | −1942.8 | <0.0001a |
| X 23 | −3847.7 | <0.0001a |
| X 24 | −377.8 | <0.0001a |
| X 25 | −334.8 | <0.0001a |
| X26 | −32.1 | <0.0001a |
| X27 | −0.1 | <0.0001a |
aSignificant at p ≤ 0.05
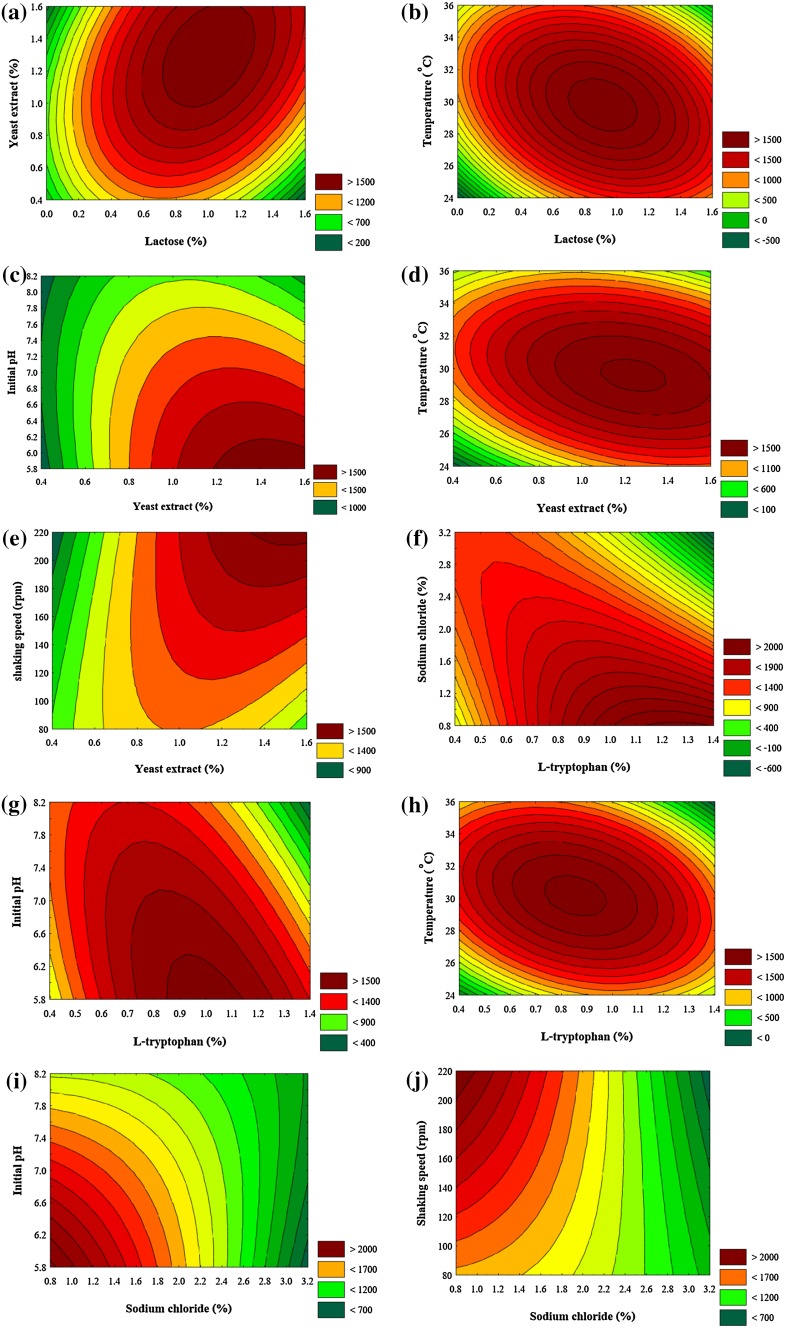
A contour plot was drawn to illustrate the interactions between the variables with regard to IAA yield. The contour plot of significant interaction terms, i.e. X 1 X 2, X 1 X 6, X 2 X 5, X 2 X 6, X 2 X 7, X 3 X 4, X 3 X 5, X 3 X 6, X 4 X 5, and X 4 X 7 is shown in Fig. 4. This analysis reveals that a high level of yeast extract, l-tryptophan, and shaking speed, medium levels of lactose and temperature, and low levels of NaCl and pH supported the production of IAA in Enterobacter sp. DMKU-RP206. The model reveals that l-tryptophan (X 3) is significantly important with regard to IAA production as indicated by the highest coefficient, followed by yeast extract (X 2) and lactose (X 1) (Table 3).
Fig. 4.
Contour plot of significant interaction-terms on IAA production by Enterobacter sp. DMKU-RP206
Validation of the experimental model was confirmed when the influencing factors were kept at 0.85% (lactose), 1.3% (yeast extract), 1.1% (l-tryptophan), 0.4% (NaCl), 5.8 (initial pH), 30 °C (temperature), and 200 rpm (shaking speed). Under these conditions, the observed and predicted values of IAA were 3804.2 and 4334.2 mg l−1, respectively. The percentage relative error between the observed and predicted values was 12.2%.
IAA production in a 2 l stirred tank fermenter
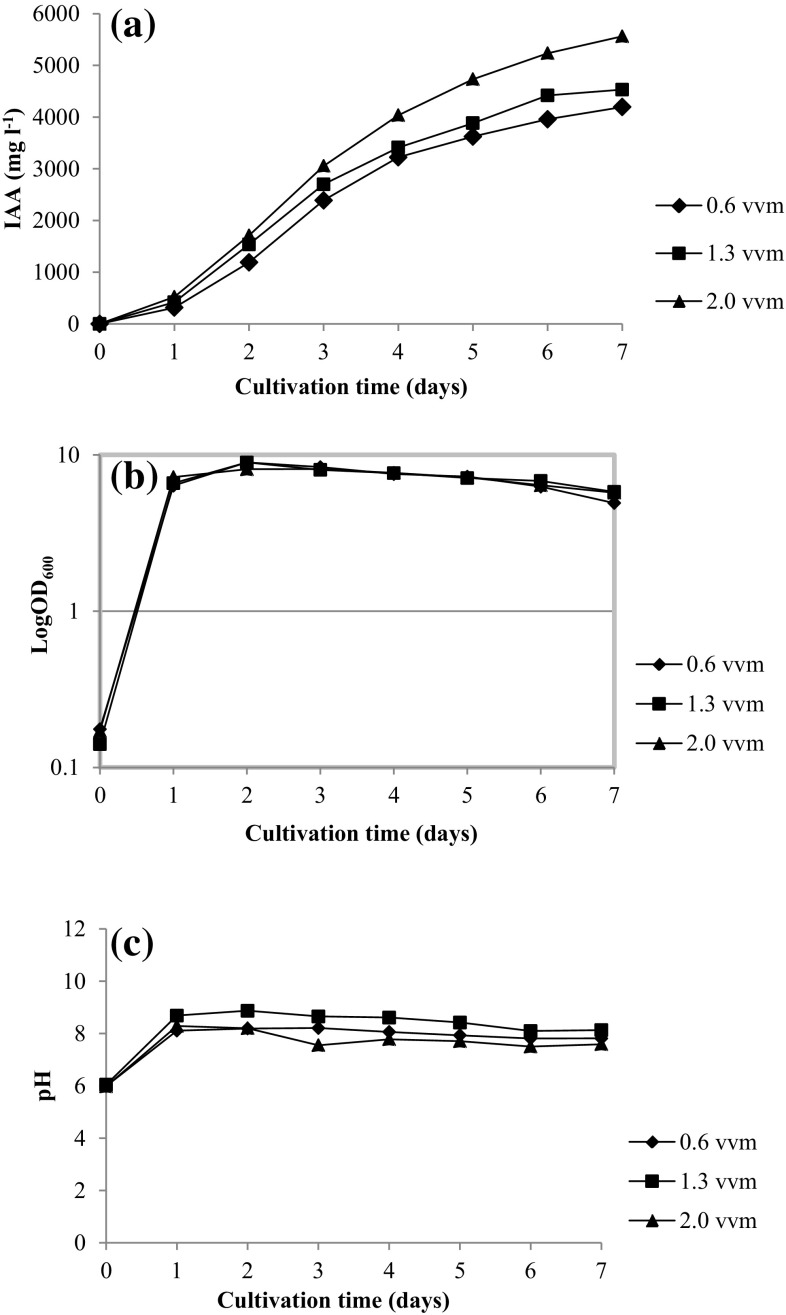
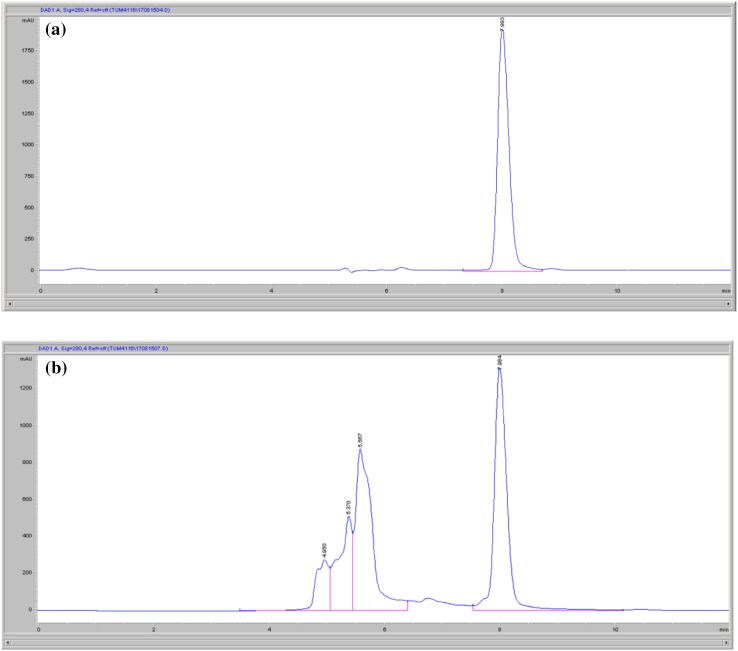
The highest IAA amount (5561.7 mg l−1) was obtained when Enterobacter sp. DMKU-RP206 was cultivated at an aeration rate of 2 vvm after 7 days of incubation. The IAA production at aeration rates of 1.3 and 0.6 vvm were 4530.7 and 4194.7 mg l−1, respectively (Fig. 5a). Regarding bacterial growth, the various cultivation conditions showed no significant difference in growth rates (Fig. 5b). Although the pH was uncontrolled, slight pH changes could be observed throughout the course of cultivation (Fig. 5c). The HPLC chromatogram of IAA in the culture supernatant is shown in Fig. 6.
Fig. 5.
IAA (a), growth (b), and pH (c) during batch fermentation in 2 l stirred tank fermenter by Enterobacter sp. DMKU-RP206 with agitation speed of 200 rpm and varied aeration rates (0.6, 1.3, and 2.0 vvm)
Fig. 6.
HPLC chromatogram of authentic indole-3-acetic acid (a) and culture supernatant of Enterobacter sp. DMKU-RP206 (b)
Discussion
Agriculture is one of the human activities that contribute the most to the increasing amount of chemical pollutants via excessive use of synthetic chemical fertilizers and pesticides. Presently, the use of microorganisms instead of synthetic chemical fertilizers and pesticides to support plant growth through direct and indirect mechanisms is being widely investigated. The benefit of microorganisms, known as plant growth promoting bacteria (PGPB), plays an important role in enhancing plant growth through a wide variety of mechanisms including (1) facilitating nutrient acquisition through nitrogen fixation and phosphate solubilization, (2) phytohormones production, (3) siderophores production, and (4) the production of pathogen cell wall-degrading enzymes to prevent plant pathogens.
Nitrogen and phosphorus are major organic compounds that improve the growth of plants. Nitrogen and phosphorus are required for plant growth and increased productivity, as well as for formation of an integral part of proteins, nucleic acids, and other essential biomolecules (Bockman 1997). Enterobacter sp. DMKU-RP206 showed ammonia production and phosphate solubilization abilities that will provide nitrogen and phosphorus to support plant growth. Similarly, iron is an essential micronutrient for plant and fungal pathogen growth as it is involved in various important biological processes, such as being a cofactor of various metabolic enzymes, photosynthesis, respiration, and chlorophyll biosynthesis (Vessey 2003; Kobayashi and Nishizawa 2012). Enterobacter sp. DMKU-RP206 also showed a capacity for siderophore production that traps iron to directly support plant growth. In addition, siderophore can also have a negative effect on fungal pathogens through iron depletion. In terms of antagonistic activity, Enterobacter sp. DMKU-RP206 showed a significant antagonistic effect against all fungi tested—i.e. Curvularia lunata, Fusarium moniliforme, and Rhizoctonia solani—which cause disease in rice plants. In a similar previous study, Enterobacter spp.—i.e. Enterobacter asburiae (Abraham and Silambarasan 2015), Enterobacter cancerogenus (Jha et al. 2012), and Enterobacter cloacae (Abraham and Silambarasan 2015; Bose et al. 2016)—support plant growth and is considered to be a plant growth promoting bacteria. Furthermore, Enterobacter sp. DMKU-RP206 exhibited an ability to produce a high level of IAA, which is one of the main direct mechanisms to promote plant growth. IAA affects plant cell division, extension, and differentiation, stimulates seed germination, initiates lateral, adventitious root formation, and responses to light and gravity (Teale et al. 2006).
Plant growth-promoting bacteria (PGPB) have been studied and some of them have already been commercialized such as Bacillus spp., Pseudomonas spp., Rhizobia spp., and Streptomyces spp. However, the utilization of PGPB in agriculture represents only a small fraction of current worldwide agricultural practice (Glick 2012). The successful application of PGPB in the field has been limited by variance of ecological factors that determine their survival and activity in the field (Martínez-Viveros et al. 2010). These disadvantages limit the application of PGPB. Therefore, this study was interested in the use of PGPB’s product, in particular IAA, instead of the PGPB cell.
Regarding IAA production, this bacterium showed a higher level of IAA when compared to previous reports on IAA production by Enterobacter (Inui-Kishi et al. 2012; Ghosh et al. 2015; Bose et al. 2016). IAA production by Enterobacter sp. DMKU-RP206 commenced after 1 day of incubation (282.4 mg IAA l−1) and reached the maximum production of 448.5 mg l−1 after 10 days. However, we decided to reduce the cultivation to only 5 days (414.2 mg IAA l−1) in order to reduce the production costs. Shorter incubation periods were observed in previous reports on IAA production by other Enterobacter species, even though only a small amount could be detected. For example, Enterobacter sp. A3CK and E. cloacae A7CK rapidly produced IAA (about 15 mg l−1) after 2 h of incubation, where the maximum amounts of IAA (about 250 and 200 mg l−1 by Enterobacter sp. A3CK and E. cloacae A7CK, respectively) were obtained after an incubation time of 20 h (Ghosh et al. 2015). E. cloacae SN19 produced IAA after 6 h of incubation (about 25 mg l−1) and this increased to about 190 mg l−1 after the first day. The maximum IAA (382.2 mg l−1) was reached after 3 days of incubation (Bose et al. 2016). In addition, Inui-Kishi et al. (2012) studied IAA production at different incubation times in four plant growth-promoting rhizobacteria. They showed that Enterobacter sp. USC8 and USC7 started to accumulate IAA after 12 and 24 h of incubation, whereas other bacteria—Burkholderia sp. A5I55 and Labrys sp. A5I42—started to produce IAA at 36 and 60 h of incubation, respectively. Regarding to Enterobacter sp. USC8 and USC7, a higher yield (136.9–152.6 mg IAA l−1) and a shorter incubation time (48–60 h) were shown in the course of IAA fermentation than those were shown with Burkholderia sp. A5I55 and Labrys sp. A5I42 (17.6–58.3 mg IAA l−1, 72–96 h). It was, therefore, indicated that the genus Enterobacter tends to be potent IAA-producing bacteria. In addition, it was obviously shown that our Enterobacter sp. DMKU-RP206 possessed the highest IAA production compared to all other Enterobacter spp. previously reported.
Studies on IAA production involving bacteria suggested that optimal carbon and nitrogen sources for IAA production were bacterial strain-dependent. Shridevi and Mallaiah (2007) showed that Rhizobium strains vary in their utilization of carbon sources and the production of IAA was also shown in terms of different carbon sources such as sucrose, inositol, and glucose. Sucrose and mannitol supported the growth and IAA production in Pantoea agglomerans PVM (Apine and Jadhav 2011), and Enterobacter sp. A3CK and E. cloacae A7CK (Ghosh et al. 2015), respectively. In the present study, lactose was shown as the best carbon source for IAA production (559.6 mg l−1) by Enterobacter sp. DMKU-RP206. This result agrees with Jeyanthi and Ganesh (2013) that the best yield of 53 mg IAA l−1 of Pseudomonas fluorescence was obtained with medium-amended lactose.
In the case of nitrogen sources, yeast extract was the best nitrogen source in terms of IAA production (754.4 mg l−1) involving Enterobacter sp. DMKU-RP206. At the same time, inorganic nitrogen compounds (monosodium glutamate, ammonium sulfate, ammonium chloride, urea, potassium nitrate, and sodium nitrite) did not support IAA production in terms of this bacterium. Yeast extract is an organic nitrogen compound containing vitamins and growth factors that support bacterial growth and also increase the availability of tryptophan with regard to IAA production in bacteria. However, this result was not in accordance with early reports on Rhizobium strains that showed maximum IAA production when growth occurred with various nitrogen sources, such as inorganic nitrogen (KNO3) and organic nitrogen (casamino acid and l-glutamic acid) (Shridevi and Mallaiah 2007). Organic nitrogen compounds, such as meat extract, that contain vitamins and growth factors gave a higher production of IAA (2104 mg l−1) in P. agglumerans PVM (Apine and Jadhav 2011).
Statistical methods were used to optimize IAA production in Enterobacter sp. DMKU-RP206. The optimal media and cultivation conditions were 0.85% lactose, 1.3% yeast extract, 1.1% l-tryptophan, 0.4% NaCl, an initial pH of 5.8, 30 °C, and a shaking speed of 200 rpm. The highest amount of IAA in a shaking flask was 3804.2 mg l−1. A 9.2-fold increase in IAA production involving Enterobacter sp. DMKU-RP206 was established when the bacterium was cultivated in TSB supplemented with 0.1% l-tryptophan for 5 days. The maximum amount of IAA found in this study was higher than those previously reported for the genus Enterobacter, such as E. ludwigii BNM 0357 (about 30 mg l−1) (Shoebitz et al. 2009), E. asburiae PS2 (32 mg l−1) (Ahemad and Khan 2010), E. arachidis Ah-143 (5.6 mg l−1) (Madhaiyan et al. 2010), E. cloacae PnB 9 (282.4 mg l−1), Enterobacter sp. PnB 10 (273.6 mg l−1) (Jasim et al. 2014), Enterobacter sp. I-3 (about 200 mg l−1) (Park et al. 2015), E. hormaechei A-2 (131.4 mg l−1), and E. cloacae subsp. dissolvens AnA-10 (119.8 mg l−1) (Arraktham et al. 2016). However, the amount of IAA produced by Enterobacter sp. DMKU-RP206 was still lower than that reported with regard to Bacillus sp. (4400 mg l−1) and Rhizobium sp. (6100 mg l−1) in optimized media containing chickpea as a substrate (Sudha et al. 2012). However, these amounts were determined by colorimetric assay which showed a higher amount of IAA than those assayed by the HPLC (de‐Bashan et al. 2008). In the present study, a scale-up on IAA production was performed to increase IAA production. As a result of variation in aeration rates, the maximum IAA was obtained at an aeration rate of 2 vvm. After 5 days of cultivation, a higher IAA concentration was found in a fermenter (4733.1 mg l−1) compared with that obtained using shaking flask cultivation (3804.2 mg l−1). The highest IAA amount of 5561.7 mg l−1 was obtained after 7 days of cultivation. This means that the bacterial cultivation in a 2-l fermenter increased IAA production by up to 1.5-fold from that found in a shaking flask culture. In this study, an overall 13.4-fold improvement in IAA production was achieved. Results showed that the highest amounts of IAA were obtained in the stationary phase growth (Figs. 4a, b), similarly to the results reported by Özdal et al. (2016). This is due to the fact that IAA is a secondary metabolite where the production occurred when the growth reached the maximal level. Expressions of the key genes for IAA production in the presence of tryptophan, e.g. indole-3-pyruvate decarboxylase gene (Vande Broek et al. 1999) and tryptophan side chain oxidase gene (Oberhansli et al. 1991) increases with the cell density and reaches its maximum at the stationary phase growth.
In the present study, lactose was the best carbon source for IAA production involving Enterobacter sp. DMKU-RP206. However, commercial lactose may not be a proper substrate to use due to high production costs. It could be replaced by an industrial dairy waste such as cheese whey. Similarly, commercial grade yeast extract may be replaced by a low-grade yeast extract. In addition, temperature adaptation to increase the Enterobacter sp. DMKU-RP206 growth temperature may be an alternative task that may reduce the cost of the cooling system for high-temperature fermentation. Moreover, the reduction of fermentation time is also an important goal. Currently, continuous fermentation of immobilized cells has been used to shorten fermentation time and increase IAA concentration as well as productivity (Özdal et al. 2016; Ozdal et al. 2017).
Conclusions
The overall results suggest that Enterobacter sp. DMKU-RP206 has great potential for IAA production in bacteria and, in particular, in terms of the genus Enterobacter. Optimization by statistical methods improves IAA production involving Enterobacter sp. DMKU-RP206. In terms of shaking flask cultivation, the maximum amount of IAA (3804.2 mg l−1) was obtained in a culture medium containing 0.85% lactose, 1.3% yeast extract, 1.1% l-tryptophan, 0.4% NaCl, pH adjusted to 5.8, and incubated on an orbital shaker at 200 rpm and 30 °C. In terms of 2 l fermenter cultivation, the highest IAA production (5561.7 mg l−1) was obtained when an aeration rate of 2 vvm and agitation speed of 200 rpm were applied. Apart from IAA production, Enterobacter sp. DMKU-RP206 can also support plant growth by various mechanisms such as direct mechanisms (phosphate solubilization and ammonia production) and indirect mechanisms (siderophore production and biocontrol agent).
Acknowledgement
This work was supported by the Higher Education Research Promotion and National Research University Project of Thailand, Office of the Higher Education Commission.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
References
- Abraham J, Silambarasan S. Plant growth promoting bacteria Enterobacter asburiae JAS5 and Enterobacter cloacae JAS7 in mineralization of endosulfan. Appl Biochem Biotechnol. 2015;175:3336–3348. doi: 10.1007/s12010-015-1504-7. [DOI] [PubMed] [Google Scholar]
- Ahemad M, Khan MS. Plant growth promoting activities of phosphatesolubilizing Enterobacter asburiae as influenced by fungicides. EurAsia J BioSci. 2010;4:88–95. doi: 10.5053/ejobios.2010.4.0.11. [DOI] [Google Scholar]
- Apine OA, Jadhav JP. Optimization of medium for indole-3-acetic acid production using Pantoea agglomerans strain PVM. J Appl Microbiol. 2011;110:1235–1244. doi: 10.1111/j.1365-2672.2011.04976.x. [DOI] [PubMed] [Google Scholar]
- Arraktham S, Tancho A, Niamsup P, Rattanawaree P. The potential of bacteria isolated from earthworm intestines, vermicompost and liquid vermicompost to produce indole-3-acetic acid (IAA) J Agric Technol. 2016;12:229–239. [Google Scholar]
- Bakker AW, Schipper B. Microbial cyanide production in the rhizosphere in relation to potato yield reduction and Pseudomonas spp. mediated plant growth stimulation. Soil Biol Biochem. 1987;19:451–457. doi: 10.1016/0038-0717(87)90037-X. [DOI] [Google Scholar]
- Bockman OC. Fertilizers and biological nitrogen fixation as sources of plant nutrients: perspectives for future agriculture. Plant Soil. 1997;194:11–14. doi: 10.1023/A:1004212306598. [DOI] [Google Scholar]
- Bose A, Kher M, Nataraj M, Keharia H. Phytostimulatory effect of indole-3-acetic acid by Enterobacter cloacae SN19 isolated from Teramnus labialis (L. f.) Spreng rhizosphere. Biocatal Agric Biotechnol. 2016;6:128–137. [Google Scholar]
- Brady C, Cleenwerck I, Venter SN, Vancanneyt M, Swings J, Coutinho TA. Phylogeny and identification of Pantoea species associated with plants, humans and the natural environment based on multilocus sequence analysis (MLSA) Syst Appl Microbiol. 2008;31:447–460. doi: 10.1016/j.syapm.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Cappuccino JC, Sherman N. Microbiology: a laboratory manual. 3. New York: Benjamin/Cumming Pub. Co; 1992. [Google Scholar]
- Cattelan AJ, Hartel PG, Fuhrmann JJ. Screening for plant growth-promoting rhizobacteria to promote early soybean growth. Soil Sci Soc Am J. 1999;63:1670–1680. doi: 10.2136/sssaj1999.6361670x. [DOI] [Google Scholar]
- Cloete KJ, Valentine AJ, Stander MA, Blomerus LM, Botha A. Evidence of symbiosis between the soil yeast Cryptococcus laurentii and a sclerophyllous Medicinal shrub, Agathosma betulina (Berg.) pillans. Microb Ecol. 2009;57:624–632. doi: 10.1007/s00248-008-9457-9. [DOI] [PubMed] [Google Scholar]
- de Souza R, Meyer J, Schoenfeld R, da Costa PB, Passaglia LM. Characterization of plant growth-promoting bacteria associated with rice cropped in iron-stressed soils. Ann Microbiol. 2015;65:951–964. doi: 10.1007/s13213-014-0939-3. [DOI] [Google Scholar]
- de-Bashan LE, Antoun H, Bashan Y. Involvement of indole-3-acetic acid produced by the growth-promoting bacterium Azospirillium spp. in promoting growth of Chlorella vulgaris. J Phycol. 2008;44:938–947. doi: 10.1111/j.1529-8817.2008.00533.x. [DOI] [PubMed] [Google Scholar]
- Dell’Amico E, Cavalca L, Andreoni V. Analysis of rhizobacterial communitiesin perennial Graminaceae from polluted water meadow soil, and screening of metal resistant, potentially plant growth-promoting bacteria. FEMS Microbiol Ecol. 2005;52:153–162. doi: 10.1016/j.femsec.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Duan YQ, Zhou XK, Di-Yan L, Li QQ, Dang LZ, Zhang YG, Qiu LH, Nimaichand S, Li WJ. Enterobacter tabaci sp. nov., a novel member of the genus Enterobacter isolated from a tobacco stem. Antonie Van Leeuwenhoek. 2015;108:1161–1169. doi: 10.1007/s10482-015-0569-1. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fu SF, Sun PF, Lu HY, Wei JY, Xiao HS, Fang WT, Cheng BY, Chou JY. Plant growth-promoting traits of yeasts isolated from the phyllosphere and rhizosphere of Drosera spatulata Lab. Fungal Biol. 2016;120:433–448. doi: 10.1016/j.funbio.2015.12.006. [DOI] [PubMed] [Google Scholar]
- Ghosh PK, Sen SK, Maiti TK. Production and metabolism of IAA by Enterobacter spp. (Gammaproteobacteria) isolated from root nodules of a legume Abrus precatorius L. Biocatal Agric Biotechnol. 2015;4:296–303. [Google Scholar]
- Glick BR. Plant growth-promoting bacteria: mechanisms and applications. Scientifica. 2012 doi: 10.6064/2012/963401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickmann E, Dessaux Y. A critical examination of the specificity of the salkowski reagent for indolic compounds produced by phytopathogenic bacteria. Appl Environ Microbiol. 1994;61:793–796. doi: 10.1128/aem.61.2.793-796.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inácio J, Pereira P, Carvalho M, Fonseca A, Amaral-Collaço MT, Spencer-Martins I. Estimation and diversity of phylloplane mycobiota on selected plants in a Mediterranean-type ecosystem in Portugal. Microb Ecol. 2002;44:344–353. doi: 10.1007/s00248-002-2022-z. [DOI] [PubMed] [Google Scholar]
- Inui-Kishi RN, Kishi LT, Picchi SC, Barbosa JC, Lemos MTO, Marcondes J, Lemos EGDM. Phosphorus solubilizing and IAA production activities in plant growth promoting rhizobacteria from brazilian soils under sugarcane cultivation. J Eng Appl Sci. 2012;7:1446–1454. [Google Scholar]
- Jasim B, John CJ, Shimil V, Jyothis M, Radhakrishnan EK. Studies on the factors modulating indole-3-acetic acid production in endophytic bacterial isolates from Piper nigrum and molecular analysis of ipdc gene. J Appl Microbiol. 2014;117:786–799. doi: 10.1111/jam.12569. [DOI] [PubMed] [Google Scholar]
- Jeyanthi V, Ganesh P. Production, optimization and characterization of phytohormone indole acetic acid by Pseudomonas fluorescence. Int J Pharm Biol Arch. 2013;4:514–520. [Google Scholar]
- Jha CK, Patel B, Saraf M. Stimulation of the growth of Jatropha curcas by the plant growth promoting bacterium Enterobacter cancerogenus MSA2. World J Microbiol Biotechnol. 2012;28:891–899. doi: 10.1007/s11274-011-0886-0. [DOI] [PubMed] [Google Scholar]
- Kämpfer P, Ruppel S, Remus R. Enterobacter radicincitans sp. nov., a plant growth promoting species of the family Enterobacteriaceae. Syst Appl Microbiol. 2005;28:213–221. doi: 10.1016/j.syapm.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Kämpfer P, McInroy JA, Glaeser SP. Enterobacter muelleri sp. nov., isolated from the rhizosphere of Zea mays. Int J Syst Evol Microbiol. 2015;65:4093–4099. doi: 10.1099/ijsem.0.000547. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Nishizawa NK. Iron uptake, translocation, and regulation in higher plants. Annu Rev Plant Biol. 2012;63:131–152. doi: 10.1146/annurev-arplant-042811-105522. [DOI] [PubMed] [Google Scholar]
- Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematic. Chichester: John Wiley and Sons; 1991. pp. 115–175. [Google Scholar]
- Madhaiyan M, Poonguzhali S, Lee JS, Saravanan VS, Lee KC, Santhanakrishnan P. Enterobacter arachidis sp. nov., a plant-growth promoting diazotrophic bacterium isolated from rhizosphere soil of groundnut. Int J Syst Evol Microbiol. 2010;60:1559–1564. doi: 10.1099/ijs.0.013664-0. [DOI] [PubMed] [Google Scholar]
- Martínez-Viveros O, Jorquera MA, Crowley DE, Gajardo GMLM, Mora ML. Mechanisms and practical considerations involved in plant growth promotion by rhizobacteria. J Soil Sci Plant Nutr. 2010;10:293–319. doi: 10.4067/S0718-95162010000100006. [DOI] [Google Scholar]
- Matsukawa E, Nakagawa Y, Iimura Y, Hayakawa M. Stimulatory effect of indole-3-acetic acid on aerial mycelium formation and antibiotic production in Streptomyces spp. Actinomycetologica. 2007;21:32–39. doi: 10.3209/saj.SAJ210105. [DOI] [Google Scholar]
- Nadeem SM, Ahmad M, Naveed M, Imran M, Zahir ZA, Crowley DE. Relationship between in vitro characterization and comparative efficacy of plant growth-promoting rhizobacteria for improving cucumber salt tolerance. Arch Microbiol. 2016;198:379–387. doi: 10.1007/s00203-016-1197-5. [DOI] [PubMed] [Google Scholar]
- Nutaratat P, Srisuk N, Arunrattiyakorn P, Limtong S. Plant growth-promoting traits of epiphytic and endophytic yeasts isolated from rice and sugar cane leaves in Thailand. Fungal Biol. 2014;118:683–694. doi: 10.1016/j.funbio.2014.04.010. [DOI] [PubMed] [Google Scholar]
- Oberhansli T, Defago G, Haas D. Indole-3-acetic-acid (IAA) synthesis in the biocontrol strain CHA0 of Pseudomonas fluorescens: role of tryptophan side chain oxidase. J Gen Microbiol. 1991;137:2273–2279. doi: 10.1099/00221287-137-10-2273. [DOI] [PubMed] [Google Scholar]
- Ozdal M, Ozdal OG, Sezen A, Algur OF, Kurbanoglu EB. Continuous production of indole-3-acetic acid by immobilized cells of Arthrobacter agilis. 3. Biotech. 2017;7:23. doi: 10.1007/s13205-017-0605-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özdal M, Özdal ÖG, Sezen A, Algur ÖF. Biosynthesis of indole-3-acetic acid by Bacillus cereus immobilized cells. Cumhuriyet Science Journal. 2016;37:212–222. doi: 10.17776/csj.34085. [DOI] [Google Scholar]
- Park JM, Radhakrishnan R, Kang SM, Lee IJ. IAA producing Enterobacter sp. I-3 as a potent bio-herbicide candidate for weed control: a special reference with lettuce growth inhibition. Indian J Microbiol. 2015;55:207–212. doi: 10.1007/s12088-015-0515-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paudel YP, Qin W. Characterization of novel cellulase-producing bacteria isolated from rotting wood samples. Appl Biochem Biotechnol. 2015;177:1186–1198. doi: 10.1007/s12010-015-1806-9. [DOI] [PubMed] [Google Scholar]
- Prusty R, Grisafi P, Fink GR. The plant hormone indole acetic acid induces invasive growth in Saccharomyces cerevisiae. Proc Natl Acad Sci. 2004;101:4153–4157. doi: 10.1073/pnas.0400659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao RP, Hunter A, Kashpur O, Normanly J. Aberrant synthesis of indole-3-acetic acid in Saccharomyces cerevisiae triggers morphogenic transition, a virulence trait of pathogenic fungi. Genetics. 2010;185:211–220. doi: 10.1534/genetics.109.112854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokhbakhsh-Zamin F, Sachdev D, Kazemi-Pour N, Engineer A, Pardesi KR, Dhakephalkar PK, Chopade BA. Characterization of plant-growth- promoting traits of Acinetobacter species isolated from rhizoshere of Pennisetum glaucum. J Microbiol Biotechnol. 2011;21:556–566. [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sato K, Kato Y, Taguchi G, Nogawa M, Yokota A, Shimosaka M. Chitiniphilus shinanonensis gen. nov., sp. nov., a novel chitin-degrading bacterium belonging to Betaproteobacteria. J Gen Appl Microbiol. 2009;55:147–153. doi: 10.2323/jgam.55.147. [DOI] [PubMed] [Google Scholar]
- Scagliola M, Pii Y, Mimmo T, Cesco S, Ricciuti P, Crecchio C. Characterization of plant growth promoting traits of bacterial isolates from the rhizosphere of barley (Hordeum vulgare L.) and tomato (Solanum lycopersicon L.) grown under Fe sufficiency and deficiency. Plant Physiol Biochem. 2016;107:187–196. doi: 10.1016/j.plaphy.2016.06.002. [DOI] [PubMed] [Google Scholar]
- Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophore. Anal Chem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- Shahid M, Hameed S, Imran A, Ali S, van Elsas JD. Root colonization and growth promotion of sunflower (Helianthus annuus L.) by phosphate solubilizing Enterobacter sp. Fs-11. World J Microbiol Biotechnol. 2012;28:2749–2758. doi: 10.1007/s11274-012-1086-2. [DOI] [PubMed] [Google Scholar]
- Shaikh S, Saraf M. Biofortification of Triticum aestivum through the inoculation of zinc solubilizing plant growth promoting rhizobacteria in field experiment. Biocatal Agric Biotechnol. 2017;9:120–126. [Google Scholar]
- Shoebitz M, Ribaudo CM, Pardo MA, Cantore ML, Ciampi L, Curá JA. Plant growth promoting properties of a strain of Enterobacter ludwigii isolated from Lolium perenne rhizosphere. Soil Biol Biochem. 2009;41:1768–1774. doi: 10.1016/j.soilbio.2007.12.031. [DOI] [Google Scholar]
- Shridevi M, Mallaiah KV. Bioproduction of indole acetic acid by Rhizobium strains isolated from root nodules of green manure crop, Sesbania sesban (L.) Merr. Iran J Biotechnol. 2007;5:178–182. [Google Scholar]
- Sudha M, Gowri RS, Prabhavathi P, Astapriya P, Devi SY, Saranya A. Production and optimization of indole acetic acid by indigenous micro flora using agro waste as substrate. Pak J Biol Sci. 2012;15:39–43. doi: 10.3923/pjbs.2012.39.43. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teale WD, Paponov IA, Palme K. Auxin in action: signaling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol. 2006;7:847–859. doi: 10.1038/nrm2020. [DOI] [PubMed] [Google Scholar]
- Tsavkelova EA, Cherdyntseva TA, Botina SG, Netrusov AI. Bacteria associated with orchid roots and microbial production of auxin. Microbiol Res. 2007;162:69–76. doi: 10.1016/j.micres.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Tsavkelova EA, Cherdyntseva TA, Klimova SY, Shestakov AI, Botina SG, Netrusov AI. Orchid-associated bacteria produce indole-3-acetic acid, promote seed germination, and increase their microbial yield in response to exogenous auxin. Arch Microbiol. 2007;188:655–664. doi: 10.1007/s00203-007-0286-x. [DOI] [PubMed] [Google Scholar]
- Vaikuntapu PR, Dutta S, Samudrala RB, Rao VRVN, Kalam S, Podite AR. Preferential promotion of Lycopersicon esculentum (tomato) growth by plant growth promoting bacteria associated with tomato. Indian J Microbiol. 2014;54:403–412. doi: 10.1007/s12088-014-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vande Broek A, Lambrecht M, Eggermont K, Vanderleyden J. Auxins upregulate expression of the indole-3-pyruvate decarboxylase gene in Azospirillum brasilense. J Bacteriol. 1999;181:1338–1342. doi: 10.1128/jb.181.4.1338-1342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vessey JK. Plant growth promoting rhizobacteria as biofertilizers. Plant Soil. 2003;255:571–586. doi: 10.1023/A:1026037216893. [DOI] [Google Scholar]
- Yamamoto S, Harayama S. PCR amplification and direct sequencing of gyrB genes with universal primers and their application to the detection and taxonomic analysis of Pseudomonas putida strains. Appl Environ Microbiol. 1995;61:1104–1109. doi: 10.1128/aem.61.3.1104-1109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J. Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol. 2017 doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi S, Usmani S, Singh BR, Musarrat J. Significance of Bacillus subtilis strain SJ-101 as a bioinoculant for concurrent plant growth promotion and nickel accumulation in Brassica juncea. Chemosphere. 2006;64:991–997. doi: 10.1016/j.chemosphere.2005.12.057. [DOI] [PubMed] [Google Scholar]