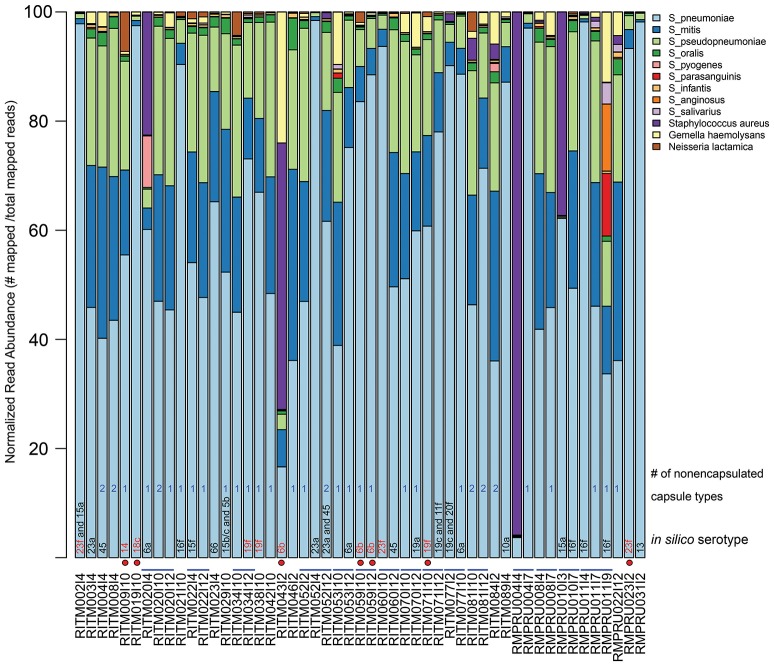
Figure 1.
Relative abundance of NP microbiome taxa from metagenomic analysis of enrichment cultures. Abundance is based on normalized read counts mapped to a reference database of Streptoccocus species and other taxa detected in the NP enrichment assemblies (Table S1). The sample names that indicate infant and sampling time point are provided under the x-axis. Blue lines connecting sample names highlight longitudinal samples originating from the same infant. In silico serotype classification was assigned using a BLAST-based strategy by aligning metagenomic assemblies against a reference database of capsule biosynthetic loci (see Methods for details). Red-colored serotype text indicates a vaccine-type serotype while a red circle depicts which vaccine-type serotype samples came from vaccinated infants. A count of the number of contigs aligning to the nonecapsulated NT_110_58-like capsule locus is given (see text for details).