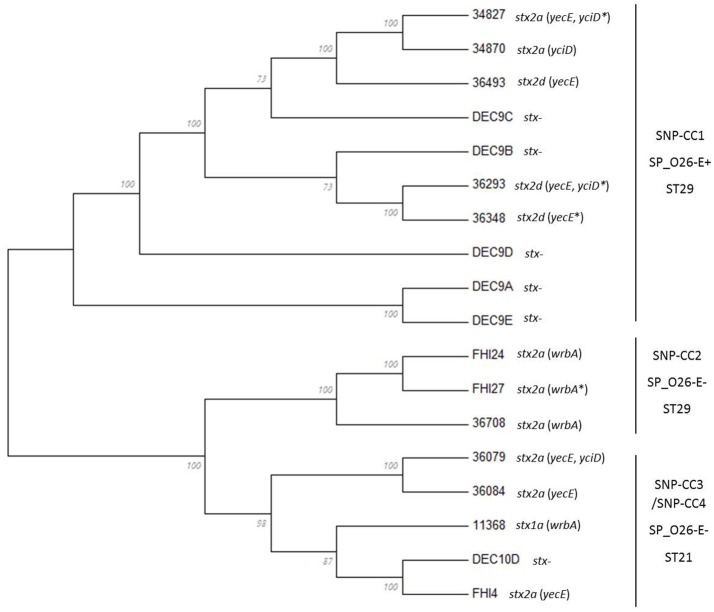
Figure 3.
Phylogenetic relationships of O26:H11 strains. The phylogenetic relationships of the 18 strains were assessed by whole-genome SNPs (wgSNP) analysis using CSI Phylogeny 1.2 on the CGE server. The SNPs alignment generated was imported and analyzed in Mega6. The evolutionary history was inferred using the Maximum Likelihood method based on the Jukes-Cantor model. The bootstrap consensus tree inferred from 100 replicates is displayed. Branches corresponding to partitions reproduced in <50% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (100 replicates) is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. All positions containing gaps and missing data were eliminated. There were a total of 4,419 positions in the final dataset. The stx subtype and insertion site(s) of the stx-phage are indicated next to the isolate name. *Indicates that the insertion site was inferred.