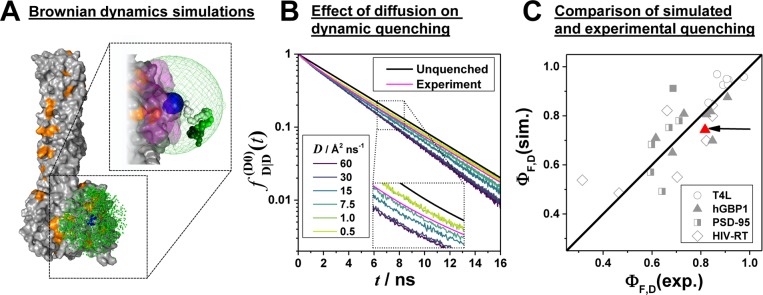
Figure 8.
Coarse-grained model captures the diffusion and dynamic quenching of Alexa488 and correlates with experimental data. Simulation of donor fluorescence decays by Brownian dynamics (BD) simulations: (A) BD simulation of the donor, D, Alexa488-C5-maleimide attached to the human guanylate binding protein 1 (PDB-ID: 1F5N). The attachment atom (on amino acid Q18C) is shown as a blue sphere, and quenching amino acids (His, Tyr, Met, and Trp) are highlighted in orange. D states close to the surface are shown in magenta. The green dots represent a subset of potential fluorophore positions of an 8 μs BD simulation. In the upper-right corner a contiguous part of a trajectory is displayed (colored from white to dark green). (B) Comparison of simulated donor fluorescence decays for various diffusion coefficients D. The analysis result of the corresponding experimental fluorescence decay, formally analyzed by a biexponential relaxation model (x1 = 0.82, τ1 = 4.15, x2 = 0.18, τ2 = 1.35), is shown in magenta. The decay of the unquenched dye with a fluorescence lifetime of 4.1 ns is shown in black. (C) Simulated fluorescence quantum yields of fluorescent species ΦF,D(sim) for a diffusion coefficient D = 15 Å/ns vs experimentally determined quantum yields ΦF,D(exp) for a set of variants of the proteins T4L, hGBP1, PSD-95, and HIV-RT. The black line shows a 1:1 relationship. ΦF,D(exp) was determined by ensemble TCSPC (hGBP1, T4L, PSD-95) or single-molecule measurements (HIV-RT). The data point highlighted by the red arrow corresponds to the experiment shown in panel B. The crystal structures used to simulate the donor fluorescence decays are listed in Table S3.