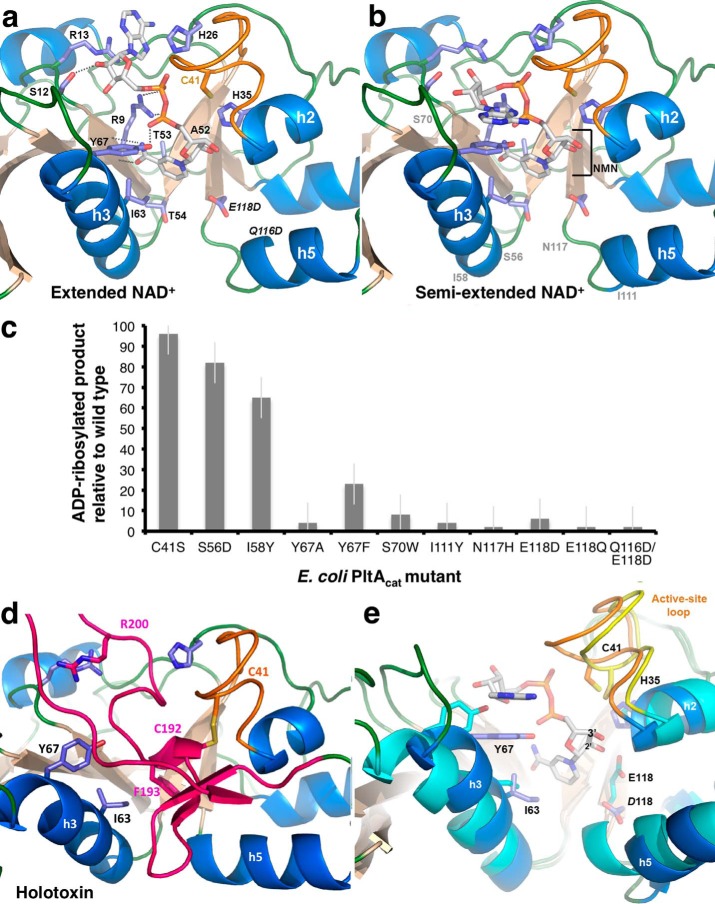
Figure 7.
NAD+-binding site of EcPltA. a, structure of EcPltA with NAD+ bound in the extended conformation; residues investigated for their role in binding and catalysis are labeled, as are the nicotinamide STS-clasp equivalent residues 52ATT54. b, structure of the semi-extended NAD+ conformation, approximate position of occluding residue mutants are shown in gray. c, ADP-ribosyltransferase assay response relative to wild-type protein for various point mutants within the NAD+-binding site. Error bars are the S.D. of three independent replicates. d, depiction of where the residues important for NAD+ binding are located in the oxidized holotoxin and their interactions with the occluding A2 helix (pink). e, overlay highlighting the structural changes occurring during activation/NAD+ binding. The semi-extended NAD+-bound structure (blue helices and orange loop) is compared with the oxidized holotoxin (cyan helices and yellow loop).