Fig. 4. Relationship between arm/chromosome and focal SCNAs and the immune or cell cycle signature scores.
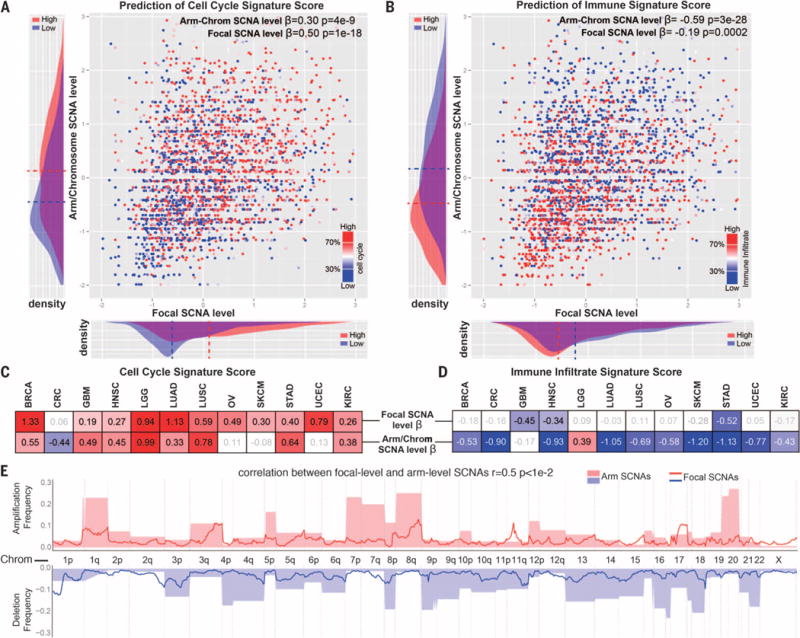
(A and B) Logistic regression was performed by comparing the tumor samples with high versus low cell cycle (A) or immune (B) signature scores and considering the arm/chromosome SCNA level and the focal SCNA level as predictors (both after standardization). The β coefficient and P value are indicated for each of the predictors. In addition, next to each of the axes on the graphs, the distribution of the corresponding parameter among tumors with high or low cell cycle (A) or immune infiltrate (B) signature is shown (dashed line indicates average value). (C and D) Logistic regression was performed as in (A) and (B) for individual tumor types. The β coefficient is indicated for each of the predictors for the indicated tumor type. A heat map is used to show the value of the β coefficients (for the coefficients with a P value of <0.1; see also table S5C). (E) Relationship between the focal and arm-level SCNAs. For each genome segment (807 genomic regions corresponding to cytogenetic bands), the frequency of amplification or deletion, for both focal and arm-level events in the pan-cancer data set, is shown (see Materials and methods and table S5D).
