Figure 1.
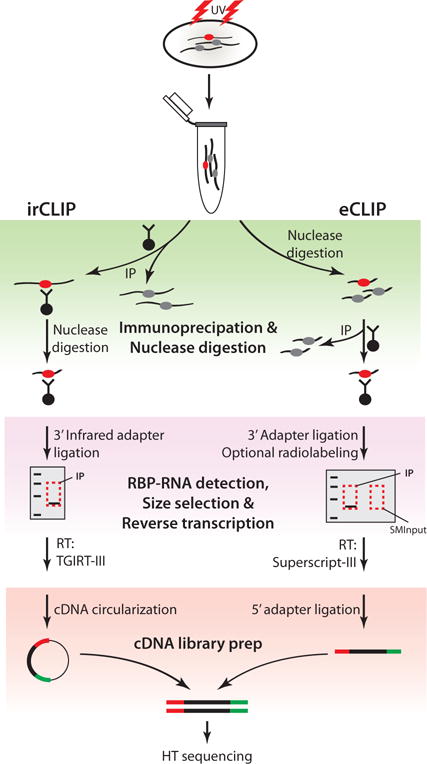
Schematic of CLIP-Seq workflow and major modifications introduced in irCLIP and eCLIP (Zarnegar et al. 2016; Van Nostrand et al. 2016). Top, cells are UV-irradiated to covalently link RNA-protein complexes, followed by lysis and RBP immunopurification. In-lysate nuclease digestion precedes RBP purification in eCLIP, while on-bead nuclease digestion is performed on immunopurified complexes in irCLIP. Ligation of an IR dye-labeled 3′ adapter in irCLIP allows rapid detection of RNA-protein complexes, rather than immunoblotting or radiolabeling as in eCLIP. Purified material is resolved by SDS-PAGE, transferred to nitrocellulose membranes, and size-selected in both methods, but eCLIP introduces purification and sequencing of size-matched RNA to allow normalization to input RNA levels (SMInput). In irCLIP, TGIRT-III reverse transcriptase is used to enhance cDNA synthesis, and a second adapter-ligation step is omitted by circularization of cDNA. In contrast, cDNA generated in eCLIP is ligated to a second adapter using optimized ligation methods. The products are then amplified by PCR and analyzed by high-throughput sequencing.
