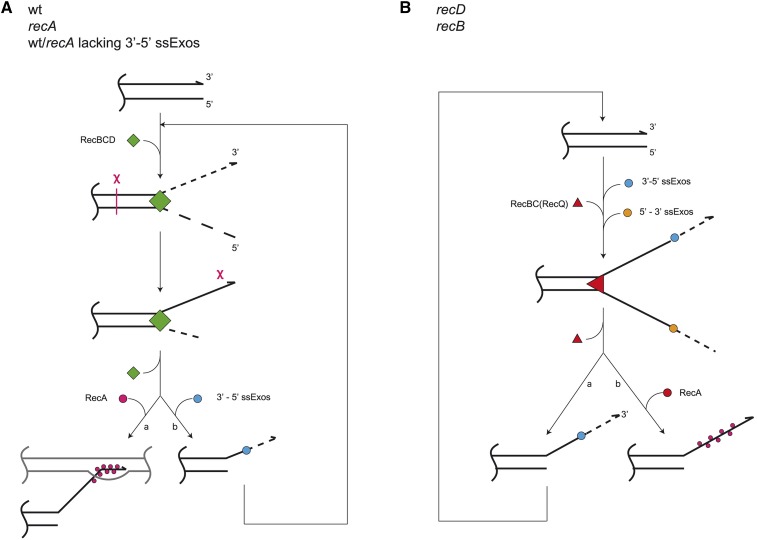
Figure 8.
Processing of a dsDNA end in several E. coli genetic backgrounds. (A) In wt bacteria, dsDNA processing is initiated by binding of RecBCD enzyme onto it, which unwinds the DNA duplex while degrading both unwound strands. Upon interaction with the χ site, the enzyme stops the 3′ strand degradation, while continuing DNA unwinding and degradation of the 5′ strand. χ-modified RecBCD also facilitates RecA loading onto a post-χ 3′ overhang (a). In the recA null mutant, RecBCD degrades both unwound strands before eventually being released from DNA, which might be independent of χ, leaving a protruding 3′ tail. The 3′ tail prevents subsequent reloading of RecBCD onto a processed dsDNA end unless/until 3′–5′ ssExos degrade it (b). Since both RecA+ and RecA− cells have similar, low DNA degradation when 3′–5′ ssExos are inactive [blocked (b) path], these exonucleases apparently act on a post-χ 3′ tail in wt cells [hence path (b) may apply to wt cells too]. (B) In recA derivatives of recB and recD mutants, dsDNA end processing starts with RecQ and RecBC helicase unwinding of the DNA duplex, respectively. The unwound 3′- and 5′-terminated strands are degraded by 3′–5′ and 5′–3′ ssExos, respectively. The degradation of the 5′ strand is apparently faster than that of its complementary 3′ strand (otherwise recD mutant would not be HR proficient) and thus the latter remains after helicase detachment from DNA. The protruding 3′ tail is then degraded by 3′–5′ ssExos (a), therefore enabling creation of a (nearly) blunt DNA end and hence repeated loading of RecQ or RecBC helicase. In RecA+ variants of recB and recD, RecA binds to the 3′ tail (b) and prevents RecQ reloading onto the DNA end in the recB mutant, while blocking its trimming and recreation of the blunt DNA end in the recD mutant and hence inhibits RecBC reloading. dsDNA, double-stranded DNA; ssExos, exonucleases that degrade a 3′ tail; wt, wild-type.