Abstract
Genetic disorders affecting the skin, genodermatoses, constitute a large and heterogeneous group of diseases, for which treatment is generally limited to management of symptoms. RNA-based therapies are emerging as a powerful tool to treat genodermatoses. In this review, we discuss in detail RNA splicing modulation by antisense oligonucleotides and RNA trans-splicing, transcript replacement and genome editing by in vitro-transcribed mRNAs, and gene knockdown by small interfering RNA and antisense oligonucleotides. We present the current state of these therapeutic approaches and critically discuss their opportunities, limitations and the challenges that remain to be solved. The aim of this review was to set the stage for the development of new and better therapies to improve the lives of patients and families affected by a genodermatosis.
Keywords: antisense oligonucleotides, in vitro-transcribed mRNA, RNA trans-splicing, siRNA, skin
1. Introduction
Genodermatoses—the inherited disorders of the skin—comprise a group of heterogeneous diseases. They display diverse clinical manifestations such as superficial epidermal and mucosal involvement, increased photosensitivity, inherited tumorigenesis and deep dermal trauma.1–3 Studies indicate that mutations in over 500 unique genes cause disorders with a distinct skin phenotype,4 which impedes the development of common therapeutic approaches. In comparison with other diseases, the development of therapies for genodermatoses has its specific advantages and challenges. The direct accessibility of the skin, the ability to culture skin cells and the possibility to reconstitute the organ in vitro facilitate the research. On the other hand, there are challenges such as multiple organ involvement, the large area that has to be treated, the avascular epidermis and site-specific heterogeneity.5 Further, for some diseases, the in vitro organ reconstruction is too simplistic because it lacks immune cells and well-developed anchoring structures.6
Therapies targeting the cause of genetic diseases can be divided into five groups: gene replacement, genome editing, protein replacement, and cell-based and RNA-based therapies.
Classic viral vector-based gene replacement therapies aim to introduce correct cDNA copies of the defective gene into affected organs. To achieve this, these approaches utilize the specific tropisms viral vectors have for certain organs.7 The limited number of vectors efficiently targeting skin cells in vivo, poor vector transmission to the epidermis and the size of some transgenes (e.g. larger than the viral vector capacity) are limitations that impede development of gene therapy approaches for skin diseases.8,9 At present, transplantation of gene-corrected skin grafts seems to be the most promising approach.10–14 In addition, genome editing is emerging as a powerful tool for gene and cell therapy.15 The technique has already been used to successfully correct skin cells in vitro.16,17 However, the therapeutic potential of direct in vivo correction remains to be evaluated for genodermatoses and may appear to be challenging.
Systemic protein replacement therapy may be an alternative for a subset of genodermatoses and will likely be most effective for proteins naturally expressed in the dermal extracellular matrix or the dermal–epidermal junction.18 Epidermal delivery can be achieved by direct application onto wounds or by microneedle injections.19,20
Several genodermatoses caused by primary immunodeficiencies can be managed by HLA-matched bone marrow transplantations.21 It has been shown that bone marrow transplantation induced some transient symptomatic improvement also in connective tissue genodermatoses. Nevertheless, as the studies presented no clear evidence of wild-type protein synthesis, the exact underlying mechanism remains elusive.22,23 The epidermis contains a pool of self-renewing stem cells sufficient to sustain organ homeostasis, which is evident from long-lasting mosaic patches that occur naturally in some genodermatoses.24,25 However, bone marrow transplantation, presently performed without further cell and niche manipulation, does not seem to substantially increase the number of curative stem cells in the epidermis and its long-term potential is thus uncertain. The local application of corrected stem cells, on the other hand, might be a more efficacious approach.8,26–28
RNA-based therapies can overcome some of the difficulties that accompany these cell, gene and protein replacement therapies, as well as the unique challenges posed by treating skin. In the following sections, we will introduce specific RNA-based therapies with their current and future applications for genodermatoses. Further, and importantly, we will critically discuss their advantages and limitations (summarized in Table 1). We will focus on approaches that directly utilize exogenously delivered RNA molecules. Post-transcriptional modification of RNA transcripts by RNA editing and modulation of translation of mutated mRNA transcripts by translational read-through are therapeutic options as well. However, for these approaches, the RNA is the therapeutic target rather than the therapeutic tool. To make a clear distinction between these two discrete strategies, we have therefore chosen to place a review of the latter approaches in the Supporting information (Appendix S1).
Table 1.
Summary of the present status of RNA-based therapies for the treatment of genodermatoses
| RNA-based therapy | Mechanism of action | Delivery | Advantages | Limitations | Disease | Key references |
|---|---|---|---|---|---|---|
| AON | Induced skipping of mutated exon | Systemic Intradermal Topical |
|
|
EB | Goto et al. (2006)29 Turczynski et al. (2012)30 van den Akker et al. (2009)35 |
| Trans-splicing | Exchange of an mRNA part, carrying a mutation, by its wild-type copy using the endogenous splicing machinery | Ex vivo (correction of skin cells and regrafting) Intraepidermal / Intradermal |
|
|
EB | Koller et al. (2014) 55 Murauer et al. (2013)50 |
| IVT mRNA-mediated transcript replacement therapy | Introduction of corrected mRNA transcripts to drive expression of wild-type proteins in vivo | Systemic Intradermal (Topical) |
|
|
- | Kormann et al. (2011)68 |
| IVT mRNA-mediated gene editing | In situ gene repair with IVT mRNA coding for optimized zinc finger, transcription activatorlike, or CRISPR-Cas9 nucleases together with a DNA template | Ex vivo (correction of skin cells and regrafting) Systemic Intradermal |
|
|
EB | Osborn et al. (2013)70 Mahiny et al. (2015)71 |
| Mutant gene/allele knockdown | Targeted degradation of mutant transcripts | Intradermal |
|
|
Pachyonychia congenita, epidermolytic palmoplantar keratoderma, EB | Atkinson et al. (2011)77 Leachman et al. (2010)74 Leslie Pedrioli et al. (2012)75 Pendaries et al. (2012)78 |
| Stop codon read-through | Induced read-through of PTCs | Oral |
|
|
Xeroderma pigmentosum, EB | Cogan et al. (2014)89 Kuschal et al. (2013)90 |
2. Splice Modulating Therapies
2.1. AON-mediated exon skipping
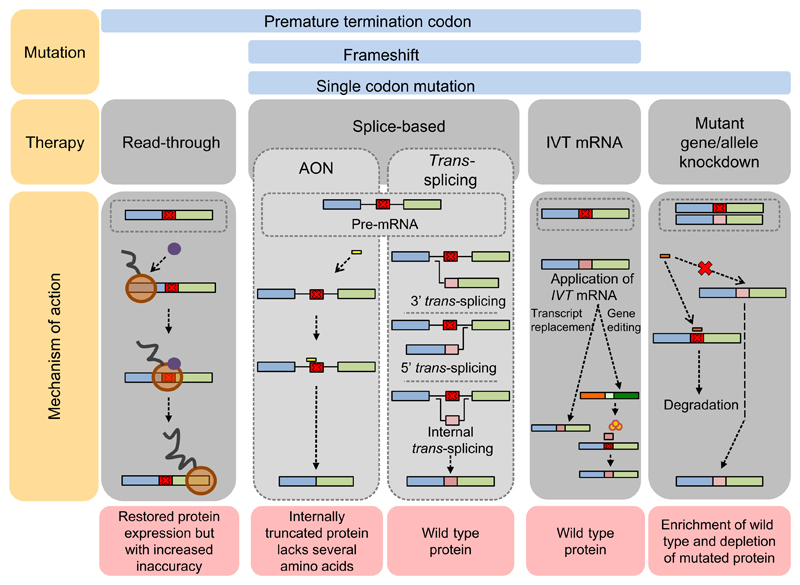
Antisense oligonucleotides (AONs) are small pieces of modified DNA or RNA. They can be exploited to, for example, knock down gene expression or to manipulate splicing. For the latter, AONs hybridize to a mutated, in-frame exon during pre-mRNA splicing (Fig. 1 and Fig. S1). The targeted exon is no longer recognized by the splicing machinery and is skipped from the mature transcript. Subsequent translation of the generated transcript will result in an internally deleted protein.
Figure 1.
RNA-based therapy approaches for genodermatoses. Graphic summary of principles and major mechanisms of the RNA-based therapy strategies presented in this review. More detail on the specific approaches can be found in their specific sections in the text and in Figs S1–S5
AON-mediated splicing modulation is being pursued for dystrophic epidermolysis bullosa (DEB), which is caused by genetic deficiency of type VII collagen, encoded by COL7A1.29–31 In 2006, Goto et al.29 showed that in vitro transfection with a specific AON targeting the mutated exon 70 of COL7A1 evoked de novo type VII collagen synthesis in patient keratinocytes. Subsequently, a single injection with this AON, directly into human skin equivalents grafted onto the back of rats, partially restored type VII collagen synthesis. Although these first results were promising, the efficiency of type VII collagen restoration was low.
AON-mediated splicing modulation is currently in clinical trials for Duchenne muscular dystrophy and spinal muscular atrophy,32 and in preclinical stages for several other genetic conditions. The hope is that the experience gained from developing AON-based therapy for these diseases will help to significantly improve the exon skipping efficiency in COL7A1 to reach therapeutically relevant levels of type VII collagen synthesis.26,31,33,34 Anticipating such advances in the development of an AON therapy for DEB, we envision that AONs will become attractive for other genodermatoses caused by mutant genes with a predominant in-frame exon organization. The greatest benefit from AON treatment is foreseen for patients with severe recessive diseases, for which even a low level of restored protein synthesis might yield significant phenotypic improvement.34–36
Given that genodermatoses often affect multiple organs,4 a major advantage is that AONs can be administered systemically.37 Additional advantages are that AONs are easy to manufacture, have low toxicity and cause limited adverse events.38 Finally, AONs only restore protein synthesis in the cells that naturally express the protein, thereby avoiding concerns regarding organ and cell type specificity.
A limitation of AONs is that not all genes or mutations are suitable targets, as targeted exons need to be in-frame, and skipping should not remove amino acid sequences that are essential for protein function. Therefore, AON-based approaches need to be critically evaluated for each targeted gene, exon and even mutation. This poses obvious challenges to drug development such as performing clinical trials and generating revenue from the product, when an AON can only be used for a few patients worldwide. Furthermore, due to turnover rate of AONs, transcripts and proteins, the effect of the therapy will be transient. Therefore, continuous treatment cycles will be needed, and it is not yet known whether lifelong treatment with AONs would be tolerated and safe. Finally, the skipping efficiency, despite improvements in AON chemistry and formulation, may still remain too low for a therapeutic effect in some disorders.
2.2. RNA trans-splicing
In recent years, spliceosome-mediated RNA trans-splicing (SMaRT) has emerged as an attractive option for the repair of mutations on the mRNA level. SMaRT uses the cellular splicing machinery to recombine an endogenous target pre-mRNA containing a mutation with an exogenously delivered pre-mRNA trans-splicing molecule (RTM) coding for part of the wild-type transcript (Fig. 1 and Fig. S2). The RTM is composed of a binding domain, which confers target specificity, and a 5′, 3′ or internal wild-type coding region to replace the part of the endogenous pre-mRNA containing the disease-causing mutation. After successful recombination, a hybrid full-length wild-type mRNA is generated, and wild-type protein synthesis restored. Proof of concept has been demonstrated in cell and animal models for a variety of human genetic diseases including Alzheimer’s disease (MAPT),39 muscular dystrophy (DYSF and TTN),40,41 haemophilia A (FVIII),42 cystic fibrosis (CFTR),43 hypertrophic cardiomyopathy (MYBPC3),44 retinitis pigmentosa (RHO)45 and epidermolysis bullosa.46–49
The first preclinical proof of concept of the potential of trans-splicing came from studies in factor VIII haemophilia A knockout mice.42 Low (< 5%) recombination frequency significantly hampered the therapeutic efficiency in the earlier studies. Since then, much effort has been put into optimization of the repair molecules. Recent studies have shown that the binding domain is the crucial factor in determination of the recombination frequency.50,51 Further, a fluorescence-based RTM screening tool has been developed to optimize trans-splicing efficiency in vitro.50,52,53 Collectively, the studies have revealed that the most efficient binding domains hybridize to exon/intron junctions, thereby reducing cis-splicing due to the blockage of competitive splicing elements within the target region, and consequently enhancing trans-splicing.46,48,51
Depending on the disorder, either in situ administration54 or correction of epidermal stem cells and generation of skin grafts is conceivable. Most of the research on genodermatoses has so far focused on EB, for which RTMs have been successfully developed and optimized for preclinical studies on PLEC, KRT14, COL17A1 and COL7A1.46–48,50,55
The first correction of an EB-associated gene by RNA trans-splicing was achieved for the PLEC gene encoding the cytolinker protein plectin.49 Subsequently, proof of concept was obtained for repair of COL7A1.48 Replacement of a 3.3-kb 3′ portion of the COL7A1 mRNA harbouring a pathogenic mutation led to restoration of full-length type VII collagen synthesis in cultured primary keratinocytes from a patient suffering from recessive DEB (RDEB). Further, when the corrected keratinocytes were used to generate skin equivalents that were subsequently transplanted onto mice, formation of type VII collagen-composed rudimentary anchoring fibrils was observed.48 Proof of concept has also been demonstrated for autosomal dominant disorders using SMaRT to exchange the first seven exons of the keratin 14 gene (KRT14), which resulted in a phenotypic reversion of patient cells in vitro.47
A major advantage of trans-splicing is that it allows a reduction in size of the transgene to be delivered (i.e. only a portion of the cDNA is used instead of the full cDNA). This in turn allows a broader range of vectors to be used, which minimizes the risk of genetic rearrangements. An additional advantage of RNA trans-splicing is that the endogenous control of gene expression is maintained.56,57 Still, however, there are challenges that have to be overcome before translation into the clinic can be realized. Concerns related to the safety of the vectors needed for RTM delivery or the RTMs themselves exhibiting unspecific trans-splicing have been raised and need to be carefully addressed. Therefore, future efforts should focus on investigating off-target events and further validating the safety of this approach. In the meantime, the most promising and safest strategy for a clinical application would be correction of stem cells followed by generation of skin grafts from single-cell clones that have undergone careful safety profiling.10 Just as with AON-mediated splicing modulation, reaching clinically relevant trans-splicing efficiency may prove to be a hurdle.
3. In Vitro-Transcribed mRNA - Based Therapy
3.1. Transcript replacement
Transcript replacement therapy introduces mRNA transcripts into cells to drive synthesis of wild-type proteins (Fig. 1 and Fig. S3).58 In situ production of a protein in cells that naturally express this protein guarantees correct post-translational modification, which helps overcome concerns about reduced functionality and immunogenicity of recombinant proteins.59 To create an active mRNA drug, a DNA template, based on wild-type or sequence-engineered cDNA,59,60 has to be transcribed in vitro and a 5′ cap and poly-A tail added61 (Fig. S3). In vitro-transcribed (IVT) mRNA can be further modified to enhance in vivo protein translation. Examples are modification of the cap and/or nucleosides to produce less immunogenic and more stable variants.61
The potential of injecting pure mRNA to restore protein synthesis and revert disease phenotypes was recognized in the early 1990s.60,62 However, due to inherent difficulties with the approach, such as rapid degradation of mRNA by RNases in the extracellular environment,63,64 immunogenicity59 and inefficient uptake of mRNA in non-immune cells,65 these studies did not lead to wider consideration of transcript replacement as a therapeutic tool. Through increased knowledge of RNA and its recognition by the host, issues with degradation and immune activation have in part been overcome.59–61,66 Nevertheless, low transfection efficiency remains a major hurdle59 and improved transfection vehicles are needed. A few different delivery approaches are conceivable to target the skin. The dermis could be targeted by systemic administration because its deeper region is highly vascularized. Another option could be intradermal injections, as spontaneous calcium-dependent uptake of mRNA occurs in dermal cells.67 Because efficient systemic targeting of the avascular epidermis will be more difficult to solve, topical application of mRNA could be an alternative option.20 Achieving cell- and tissue-specific delivery is another challenge.68
Transcript replacement therapy is still in its infancy. We here illustrate its potential by a groundbreaking preclinical study on a monogenetic disorder affecting another squamous epithelium. Surfactant protein B (SFTPB) deficiency (OMIM 178640) is an autosomal recessive condition with neonatal lethality due to respiratory failure.69 Direct administration of Sftpb mRNA to the lung under high pressure rescued conditional Sftpb knockout mice from lethality by synthesis of surfactant protein B.68 This success was achieved after optimizing both delivery of the mRNA to the lung and protein translation by mRNA modifications.68
3.2. IVT mRNA-mediated genome editing
IVT mRNA can also be used for genome editing. Here, the IVT mRNAs encode site-specific nucleases, for example zinc fingers, transcription activator-like nucleases (TALEN) or CRISPR-Cas9 that can be designed to generate double-stranded DNA breaks at specific target sites, which are determined by the genomic sequence. To permanently correct a gene, an IVT mRNA encoding such site-specific nucleases can be provided to cells together with a DNA template. After double-stranded breaks created by the nuclease, which is encoded by the IVT mRNA, the DNA template may be inserted between the breaks during repair by homologous recombination. The outcome of this approach is correction of the mutated gene and restoration of wild-type protein synthesis (Fig. 1 and Fig. S3).59 Of note, when no correct DNA template is provided, the DNA repair system will repair the breaks through non-homologous end joining.
IVT mRNA-mediated genome editing has been used to correct COL7A1 RDEB patient fibroblasts in vitro.70 Transfection with IVT mRNA encoding patient-specific TALEN promoted homology-directed repair, but with much lower efficiency than transfection with a plasmid encoding the TALEN. This indicates that mRNA-mediated gene editing is feasible in skin cells, but that further refinement is needed to make it an attractive alternative. In vivo proof of concept of the approach has been obtained from restoration of surfactant protein B synthesis in the above-mentioned conditional Sftpb knockout mouse.71
A clear advantage of in situ genome editing is that the genome itself is corrected, which would theoretically require only a limited number of treatments to obtain long-standing effects. In addition, in contrast to direct IVT mRNA delivery, cell- and tissue-specific targeting is not a prerequisite, because the natural regulation of gene expression is not changed. Furthermore, because the nuclease activity is limited in time due to the inherited instability of the transfected RNA, this is a safer option than vector-mediated transfer.59,71 Still, safety issues from both potential off-target nuclease activity and viral vector-based DNA template delivery remain a concern. A yet unexplored challenge in a future clinical phase will again be the delivery efficiency of both IVT mRNAs and DNA templates to the target cells.
4. Mutant Gene/ Allele Knockdown
Small interfering RNAs (siRNAs) are double-stranded RNA molecules (frequently 20–25 bp long) that target mRNA for degradation utilizing the endogenous RNA-induced silencing complex (RISC). They were first used in 1995 for silencing protein synthesis in vivo.72 Because siRNA may discriminate between two sequences differing by only one nucleotide, it constitutes a particularly interesting therapeutic option for dominantly inherited genodermatoses (Fig. 1 and Fig. S4). Therefore, most work has been performed on the dominantly inherited diseases such as keratinopathies (reviewed in Ref. 73) pachyonychia congenita74 (KRT6A, KRT6B, KRT16 or KRT17), epidermolytic palmoplantar keratoderma,75,76 (KRT9), and on EB simplex (KRT5, KRT14).77 A search on www.clinicaltrials.gov for “siRNA” identified one genodermatosis study among 39 registered trials. This phase Ib trial for the treatment of pachyonychia congenita was a single-patient, double-blinded, split-body, vehicle-controlled, dose-escalation trial.74 The trial evaluated the safety and efficacy of TD101, an siRNA specifically designed against the mRNA encoding the keratin 6a p.Asn171Lys mutant. Intradermal injections were performed in symmetric plantar calluses on opposite feet. Although the results were promising, with regression of the callus on the siRNA-treated side, the intense pain experienced by the patient due to the injections was a significant concern. Thus, improved topical delivery methods will be necessary. siRNAs have also been pursued for dominant DEB.78,79 The rationale there is to change the ratio between mutant and wild-type COL7A1 transcripts by depleting the mutant version. This would result in increased synthesis of wild-type type VII collagen.80
These siRNA strategies have the disadvantage of being mutation-specific. To overcome this, a mutation-independent strategy has been proposed.79 The strategy is based on administration of siRNAs to knock down endogenous COL7A1 mRNA in conjunction with a sequence-modified COL7A1 cDNA that is not suppressed by the siRNA.
AONs can also be utilized to knock down mutant mRNA transcripts. Although not yet described for genodermatoses, this approach is being evaluated for the treatment of other genetic conditions like the neurodegenerative disorder Huntington’s disease. In this disorder, AONs are used to target single nucleotide polymorphisms that are in cis with the mutation to knock down the mutant mRNA by activating RNase H-mediated degradation.81 The approach is applicable to an array of mutations and patients and could thus also be relevant to several of the genes affected in dominant genodermatoses harbouring single nucleotide polymorphisms with high minor allele frequencies. Alternatively, AONs could also be designed in a mutation-specific manner, but that comes with the same disadvantages as described in the section on exon skipping by AONs.
5. Future Perspectives and Concluding Remarks
It is clear that major advances are being made in the field of therapy development for genodermatoses. The surface area of the skin and the fact that many genodermatoses also manifest in other tissues complicate development of causal therapies. Here, the RNA modulating therapies detailed in this review may have favourable properties because they generally use smaller tools than the traditional gene and cell therapy approaches. These properties facilitate systemic delivery. For very small oligonucleotides, topical delivery might be a possibility. Despite all the advancements made, there is still much room for improvement for RNA-based therapies. To facilitate this, good animal models are required. However, there are relatively few genodermatoses animal models that can be used to study RNA modulating therapies,82 and many of the existing models are cDNA knock-ins, which cannot be used to optimize RNA modulating approaches.83 Thus, new genome DNA-based animal models are needed. Animal models might also shed light on treatment cycles. However, treatment regimens depend on many variables such as protein stability, the nature of the mutation, disease history and disease modifiers. Ultimately, treatment cycles may have to be determined for each disease, patient subset or even each individual patient independently.
For many disorders, the disease might evolve due to inflammation and organ damage, which activate secondary self-perpetuating mechanisms that are not a direct cause of the genetic defect.84 Consequently, it is foreseen that the potential benefits even of causal RNA-based therapies will largely depend on the timing of therapeutic interventions. In this regard, it is generally accepted that the therapeutic benefit is larger when patients are treated in the early stages of the disease.85 It may also be necessary to combine therapeutic approaches that restore protein function with, for instance, anti-inflammatory or antifibrotic drugs to maintain good quality of life.
Another challenge is that genodermatoses are rare diseases, and therefore, the number of patients in which therapeutic approaches can be tested is limited. This is even more complicated for the RNA modulating therapies that are mutation-specific because they are only applicable to a subset of an already small group of patients. Each therapeutic compound is considered a separate medicinal product and must be evaluated separately. When patient numbers are very small, this poses obvious challenges. In this regard, a lot can be learned from the way the DMD community is trying to solve these issues in the development of the exon skipping approach. Here, several stakeholder meetings have been organized with academic researchers, regulators, patient representatives and pharmaceutical companies to discuss the challenges and opportunities.86,87 These discussions imply that it may be possible to have smaller trials for additional DMD exon skipping AONs once clinical benefit has been convincingly shown for multiple AONs. The discussions also made clear that it is critically important to be prepared for clinical trials. They further underscored the importance of clinical trial registries for rare diseases like those coordinated by TREAT-NMD (a clinical trial infrastructure network for neuromuscular disorders, http://www.treatnmd.eu/) to facilitate selecting as many patients as possible to participate in clinical trials.88 Additionally, the insufficient understanding of the natural progression of many genodermatoses will likely impede translation of therapies into the clinic because it makes it challenging to define relevant trial endpoints. As more potential therapies are entering the clinical trial stage, there is an increasing demand for natural history studies for genodermatoses, which are now being planned and pursued (https://clinicaltrials.gov/. Accessed April 28, 2016).
The increased interest in rare diseases from newly formed disease-specific companies or large pharmaceutical companies is making the future brighter for affected individuals and their families. So where will RNA-based therapies for genodermatoses stand a decade from now? It is likely that by then a few approaches will be undergoing clinical trials. The top candidates for fast clinical implementation are therapies where studies in other diseases have paved the way, that is AONs and stop codon read-through therapies. Other approaches will likely take longer to develop, but they may, on the other hand, yield greater clinical benefit. It is therefore imperative to continue pursuing research on all varieties of RNA-based therapies.
Supplementary Material
Acknowledgements
We thank Jackie Senior, UMC Groningen and Dr. Willeke van Roon, Leiden University Medical Center, the Netherlands, for editing the manuscript and Dr Markus Mezger University of Tübingen for stimulating discussions on IVT mRNA therapy. The work was supported by a grant from the German Federal Ministry for Education and Research (BMBF), the Netherlands Organisation for Health Research and Development (ZonMW) and Austrian Science Fund (FWF; Project I1175-B13) under the frame of Erare-2, the ERA-Net for Research on Rare Diseases (SpliceEB) to AN, AMGP, AAR and EMM. Grants for the Dutch Butterfly Child Foundation (Vlinderkind), DEBRA International to AMGP, EMM and AN, and the Netherlands Organisation for Health Research and Development (ZonMW, grant 90715614) to PCvdA, respectively, also sponsored this research. AAR, JB, AMGP and PCvdA are part of COST Action BM1207 (www.exonskipping.eu).
Abbreviations
- AON
antisense oligonucleotide
- DEB
dystrophic epidermolysis bullosa
- DMD
Duchenne muscular dystrophy
- IVT mRNA
in vitro-transcribed mRNA
- PTC
premature termination codon
- RDEB
recessive dystrophic epidermolysis bullosa
- RTM
pre-mRNA trans-splicing molecule
- siRNA
small interfering RNA
Footnotes
Conflict of Interest
AAR is employed by LUMC, which holds patents on exon skipping technology for DMD. Some of the patents have been licensed to BioMarin. As co-inventor on some of the patents, AAR is entitled to a share of any royalties. AAR also acts as an ad hoc consultant for PTC Therapeutics, BioMarin, Global Guidepoint, GLC Consulting, Deerefield Consulting, Grunenthal and BioClinica and is on the SAB of ProQR and Philae Pharmaceuticals. Remuneration for these activities goes to the LUMC. JB, AMGP and PvdA are employed by UMCG, which holds patents on exon skipping for DEB. As co-inventors, JB, AMGP and PvdA are eligible to receive a share and/or royalties. AMGP has signed a statement that she will receive no share or royalties from this patent.
Author Contribution
OB, PP, JB, UK, PCvdA, AAR, AMGP, EMM and AN wrote and critically revised the review, and approved the submitted and final versions; OB and AN designed and prepared figures; AN coordinated the project.
References
- 1.Ishida-Yamamoto A, Igawa S. J Dermatol Sci. 2014;74:99–105. doi: 10.1016/j.jdermsci.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Castori M, Morrone A, Kanitakis J, Grammatico P. Eur J Dermatol. 2012;22:299–309. doi: 10.1684/ejd.2011.1633. [DOI] [PubMed] [Google Scholar]
- 3.Has C, Nystrom A. Curr Top Membr. 2015;76:117–170. doi: 10.1016/bs.ctm.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Feramisco JD, Sadreyev RI, Murray ML, Grishin NV, Tsao H. J Invest Dermatol. 2009;129:2628–2636. doi: 10.1038/jid.2009.108. [DOI] [PubMed] [Google Scholar]
- 5.Sriram G, Bigliardi PL, Bigliardi-Qi M. Eur J Cell Biol. 2015;94:483–512. doi: 10.1016/j.ejcb.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Iriyama S, Tsunenaga M, Amano S, Adachi E. Exp Dermatol. 2011;20:953–955. doi: 10.1111/j.1600-0625.2011.01347.x. [DOI] [PubMed] [Google Scholar]
- 7.Limberis MP. Acta Biochim Biophys Sin. 2012;44:632–640. doi: 10.1093/abbs/gms036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chamcheu JC, Adhami VM, Siddiqui IV, Mukhtar H. Cutaneous cell- and gene-based therapies for inherited and acquired skin disorders. In: Templeton NS, editor. Gene and Cell Therapy Therapeutic Mechanisms and Strategies. Boca Raton, Florida, USA: CRC Press; 2015. pp. 1091–1122. [Google Scholar]
- 9.Sallach J, Di Pasquale G, Larcher F, et al. Mol Ther. 2014;22:929–939. doi: 10.1038/mt.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Droz-Georget Lathion S, Rochat A, Knott G, et al. EMBO Mol Med. 2015;7:380–393. doi: 10.15252/emmm.201404353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Luca M, Pellegrini G, Mavilio F. Br J Dermatol. 2009;161:19–24. doi: 10.1111/j.1365-2133.2009.09243.x. [DOI] [PubMed] [Google Scholar]
- 12.Mavilio F, Pellegrini G, Ferrari S, et al. Nat Med. 2006;12:1397–1402. doi: 10.1038/nm1504. [DOI] [PubMed] [Google Scholar]
- 13.Cavazza A, Mavilio F. Curr Pharm Biotechnol. 2012;13:1868–1876. doi: 10.2174/138920112802273119. [DOI] [PubMed] [Google Scholar]
- 14.Chamcheu JC, Wood GS, Siddiqui IA, et al. Exp Dermatol. 2012;21:481–489. doi: 10.1111/j.1600-0625.2012.01534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maeder ML, Gersbach CA. Mol Ther. 2016;24:430–446. doi: 10.1038/mt.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Izmiryan A, Danos O, Hovnanian A. J Invest Dermatol. 2016;136:872–875. doi: 10.1016/j.jid.2015.11.028. [DOI] [PubMed] [Google Scholar]
- 17.Chamorro C, Mencia A, Almarza D, et al. Mol Ther Nucleic Acids. 2016;5:e307. doi: 10.1038/mtna.2016.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hou Y, Guey LT, Wu T, et al. J Invest Dermatol. 2015;135:3060–3067. doi: 10.1038/jid.2015.291. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, Ghasri P, Amir M, et al. Mol Ther. 2013;21:1335–1344. doi: 10.1038/mt.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCrudden MT, McAlister E, Courtenay AJ, Gonzalez-Vazquez P, Singh TR, Donnelly RF. Exp Dermatol. 2015;24:561–566. doi: 10.1111/exd.12723. [DOI] [PubMed] [Google Scholar]
- 21.Worth AJ, Booth C, Veys P. Curr Opin Hematol. 2013;20:501–508. doi: 10.1097/MOH.0b013e328365a13b. [DOI] [PubMed] [Google Scholar]
- 22.Tolar J, Blazar BR, Wagner JE. Stem Cells. 2011;29:900–906. doi: 10.1002/stem.647. [DOI] [PubMed] [Google Scholar]
- 23.Watt SA, Dayal JH, Wright S, et al. PLoS One. 2015;10:e0137639. doi: 10.1371/journal.pone.0137639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiritsi D, Garcia M, Brander R, et al. J Invest Dermatol. 2014;134:2097–2104. doi: 10.1038/jid.2014.118. [DOI] [PubMed] [Google Scholar]
- 25.Pasmooij AM, Pas HH, Deviaene FC, Nijenhuis M, Jonkman MF. Am J Hum Genet. 2005;77:727–740. doi: 10.1086/497344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuhl T, Mezger M, Hausser I, Handgretinger R, Bruckner-Tuderman L, Nystrom A. Mol Ther. 2015;23:1368–1379. doi: 10.1038/mt.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wenzel D, Bayerl J, Nystrom A, Bruckner-Tuderman L, Meixner A, Penninger JM. Sci Transl Med. 2014;6:264ra165. doi: 10.1126/scitranslmed.3010083. [DOI] [PubMed] [Google Scholar]
- 28.Umegaki-Arao N, Pasmooij AM, Itoh M, et al. Sci Transl Med. 2014;6:264ra164. doi: 10.1126/scitranslmed.3009342. [DOI] [PubMed] [Google Scholar]
- 29.Goto M, Sawamura D, Nishie W, et al. J Invest Dermatol. 2006;126:2614–2620. doi: 10.1038/sj.jid.5700435. [DOI] [PubMed] [Google Scholar]
- 30.Turczynski S, Titeux M, Pironon N, Hovnanian A. Methods Mol Biol. 2012;867:221–238. doi: 10.1007/978-1-61779-767-5_15. [DOI] [PubMed] [Google Scholar]
- 31.Turczynski S, Titeux M, Tonasso L, Hovnanian A. J Invest Dermatol. 2013;133:S144. doi: 10.1016/j.jid.2016.07.029. [DOI] [PubMed] [Google Scholar]
- 32.Kole R, Krieg AM. Adv Drug Deliv Rev. 2015;87:104–107. doi: 10.1016/j.addr.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 33.Lu QL, Cirak S, Partridge T. Mol Ther Nucleic Acids. 2014;3:e152. doi: 10.1038/mtna.2014.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwieger-Briel A, Weibel L, Chmel N, et al. Br J Dermatol. 2015;173:1308–1311. doi: 10.1111/bjd.13945. [DOI] [PubMed] [Google Scholar]
- 35.van den Akker PC, van Essen AJ, Kraak MM, et al. J Dermatol Sci. 2009;56:9–18. doi: 10.1016/j.jdermsci.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 36.Lehmann J, Schubert S, Emmert S. J Dtsch Dermatol Ges. 2014;12:867–872. doi: 10.1111/ddg.12419. [DOI] [PubMed] [Google Scholar]
- 37.van Putten M, Young C, van den Berg S, et al. Mol Ther Nucleic Acids. 2014;3:e211. doi: 10.1038/mtna.2014.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koo T, Wood MJ. Hum Gene Ther. 2013;24:479–488. doi: 10.1089/hum.2012.234. [DOI] [PubMed] [Google Scholar]
- 39.Avale ME, Rodriguez-Martin T, Gallo JM. Hum Mol Genet. 2013;22:2603–2611. doi: 10.1093/hmg/ddt108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Philippi S, Lorain S, Beley C, et al. Hum Mol Genet. 2015;24:4049–4060. doi: 10.1093/hmg/ddv141. [DOI] [PubMed] [Google Scholar]
- 41.Monjaret F, Bourg N, Suel L, et al. Mol Ther. 2014;22:1176–1187. doi: 10.1038/mt.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chao H, Mansfield SG, Bartel RC, et al. Nat Med. 2003;9:1015–1019. doi: 10.1038/nm900. [DOI] [PubMed] [Google Scholar]
- 43.Liu X, Luo M, Zhang LN, et al. Hum Gene Ther. 2005;16:1116–1123. doi: 10.1089/hum.2005.16.1116. [DOI] [PubMed] [Google Scholar]
- 44.Mearini G, Stimpel D, Kramer E, et al. Mol Ther Nucleic Acids. 2013;2:e102. doi: 10.1038/mtna.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berger A, Lorain S, Josephine C, et al. Mol Ther. 2015;23:918–930. doi: 10.1038/mt.2015.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koller U, Wally V, Mitchell LG, et al. Nucleic Acids Res. 2011;39:e108. doi: 10.1093/nar/gkr465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wally V, Brunner M, Lettner T, et al. Hum Mol Genet. 2010;19:4715–4725. doi: 10.1093/hmg/ddq405. [DOI] [PubMed] [Google Scholar]
- 48.Murauer EM, Gache Y, Gratz IK, et al. J Invest Dermatol. 2011;131:74–83. doi: 10.1038/jid.2010.249. [DOI] [PubMed] [Google Scholar]
- 49.Wally V, Klausegger A, Koller U, et al. J Invest Dermatol. 2008;128:568–574. doi: 10.1038/sj.jid.5701152. [DOI] [PubMed] [Google Scholar]
- 50.Murauer EM, Koller U, Hainzl S, Wally V, Bauer JW. Hum Gene Ther Methods. 2013;24:19–27. doi: 10.1089/hgtb.2012.180. [DOI] [PubMed] [Google Scholar]
- 51.Wally VKU, Bauer JW. Human Genetic Diseases. Rijeka: INTECH; 2011. pp. 223–240. [Google Scholar]
- 52.Koller U, Hainzl S, Kocher T, et al. Int J Mol Sci. 2015;16:1179–1191. doi: 10.3390/ijms16011179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bauer JW, Murauer EM, Wally V, Koller U. Methods Mol Biol. 2013;961:441–455. doi: 10.1007/978-1-62703-227-8_30. [DOI] [PubMed] [Google Scholar]
- 54.Peking P, Koller U, Hainzl S, et al. Mol Ther Nucleic Acids. 2016;5:e287. doi: 10.1038/mtna.2016.3. [DOI] [PubMed] [Google Scholar]
- 55.Koller U, Wally V, Bauer JW, Murauer EM. Mol Ther Nucleic Acids. 2014;3:e157. doi: 10.1038/mtna.2014.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mansfield SG, Clark RH, Puttaraju M, et al. RNA. 2003;9:1290–1297. doi: 10.1261/rna.5101903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tahara M, Pergolizzi RG, Kobayashi H, et al. Nat Med. 2004;10:835–841. doi: 10.1038/nm1086. [DOI] [PubMed] [Google Scholar]
- 58.Yamamoto A, Kormann M, Rosenecker J, Rudolph C. Eur J Pharm Biopharm. 2009;71:484–489. doi: 10.1016/j.ejpb.2008.09.016. [DOI] [PubMed] [Google Scholar]
- 59.Sahin U, Kariko K, Tureci O. Nat Rev Drug Discovery. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 60.Thess A, Grund S, Mui BL, et al. Mol Ther. 2015;23:1456–1464. doi: 10.1038/mt.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pardi N, Muramatsu H, Weissman D, Kariko K. Methods Mol Biol. 2013;969:29–42. doi: 10.1007/978-1-62703-260-5_2. [DOI] [PubMed] [Google Scholar]
- 62.Jirikowski GF, Sanna PP, Maciejewski-Lenoir D, Bloom FE. Science. 1992;255:996–998. doi: 10.1126/science.1546298. [DOI] [PubMed] [Google Scholar]
- 63.Landre JBP, Hewett PW, Olivot JM, et al. J Cell Biochem. 2002;86:540–552. doi: 10.1002/jcb.10234. [DOI] [PubMed] [Google Scholar]
- 64.Harder J, Schroder JM. J Biol Chem. 2002;277:46779–46784. doi: 10.1074/jbc.M207587200. [DOI] [PubMed] [Google Scholar]
- 65.Youn H, Chung JK. Expert Opin Biol Ther. 2015;15:1337–1348. doi: 10.1517/14712598.2015.1057563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kariko K, Buckstein M, Ni H, Weissman D. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 67.Probst J, Weide B, Scheel B, et al. Gene Ther. 2007;14:1175–1180. doi: 10.1038/sj.gt.3302964. [DOI] [PubMed] [Google Scholar]
- 68.Kormann MS, Hasenpusch G, Aneja MK, et al. Nat Biotechnol. 2011;29:154–157. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- 69.Griese M, Lorenz E, Hengst M, et al. Pediatr Res. 2016;79:34–41. doi: 10.1038/pr.2015.173. [DOI] [PubMed] [Google Scholar]
- 70.Osborn MJ, Starker CG, McElroy AN, et al. Mol Ther. 2013;21:1151–1159. doi: 10.1038/mt.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mahiny AJ, Dewerth A, Mays LE, et al. Nat Biotechnol. 2015;33:584–586. doi: 10.1038/nbt.3241. [DOI] [PubMed] [Google Scholar]
- 72.Guo S, Kemphues KJ. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- 73.Knobel M, O’Toole EA, Smith FJ. Cell Tissue Res. 2015;360:583–589. doi: 10.1007/s00441-014-2105-4. [DOI] [PubMed] [Google Scholar]
- 74.Leachman SA, Hickerson RP, Schwartz ME, et al. Mol Ther. 2010;18:442–446. doi: 10.1038/mt.2009.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leslie Pedrioli DM, Fu DJ, Gonzalez-Gonzalez E, et al. J Invest Dermatol. 2012;132:1627–1635. doi: 10.1038/jid.2012.28. [DOI] [PubMed] [Google Scholar]
- 76.Fu DJ, Thomson C, Lunny DP, et al. J Invest Dermatol. 2014;134:754–763. doi: 10.1038/jid.2013.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Atkinson SD, McGilligan VE, Liao H, et al. J Invest Dermatol. 2011;131:2079–2086. doi: 10.1038/jid.2011.169. [DOI] [PubMed] [Google Scholar]
- 78.Pendaries V, Gasc G, Titeux M, Tonasso L, Mejia JE, Hovnanian A. J Invest Dermatol. 2012;132:1741–1743. doi: 10.1038/jid.2012.11. [DOI] [PubMed] [Google Scholar]
- 79.Morgan CP, Allen DS, Millington-Ward S, O’Dwyer GE, Palfi A, Farrar GJ. J Invest Dermatol. 2013;133:2793–2796. doi: 10.1038/jid.2013.241. [DOI] [PubMed] [Google Scholar]
- 80.Fritsch A, Spassov S, Elfert S, et al. J Biol Chem. 2009;284:30248–30256. doi: 10.1074/jbc.M109.045294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kay C, Skotte NH, Southwell AL, Hayden MR. Clin Genet. 2014;86:29–36. doi: 10.1111/cge.12385. [DOI] [PubMed] [Google Scholar]
- 82.Nystrom A, Buttgereit J, Bader M, et al. PLoS One. 2013;8:e64243. doi: 10.1371/journal.pone.0064243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bruckner-Tuderman L, McGrath JA, Robinson EC, Uitto J. J Invest Dermatol. 2010;130:1485–1488. doi: 10.1038/jid.2010.75. [DOI] [PubMed] [Google Scholar]
- 84.Nystrom A, Thriene K, Mittapalli V, et al. EMBO Mol Med. 2015;7:1211–1228. doi: 10.15252/emmm.201505061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pees C, Laccone F, Hagl M, Debrauwer V, Moser E, Michel-Behnke I. Am J Cardiol. 2013;112:1477–1483. doi: 10.1016/j.amjcard.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 86.Muntoni F, Guicheney P, Voit T. Neuromuscul Disord. 2009;19:229–234. doi: 10.1016/j.nmd.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 87.Aartsma-Rus A, Ferlini A, Goemans N, et al. Hum Gene Ther. 2014;25:885–892. doi: 10.1089/hum.2014.086. [DOI] [PubMed] [Google Scholar]
- 88.Rodger S, Lochmuller H, Tassoni A, et al. Orphanet J Rare Dis. 2013;8:171. doi: 10.1186/1750-1172-8-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cogan J, Weinstein J, Wang X, et al. Mol Ther. 2014;22:1741–1752. doi: 10.1038/mt.2014.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kuschal C, DiGiovanna JJ, Khan SG, Gatti RA, Kraemer KH. Proc Natl Acad Sci U S A. 2013;110:19483–19488. doi: 10.1073/pnas.1312088110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.