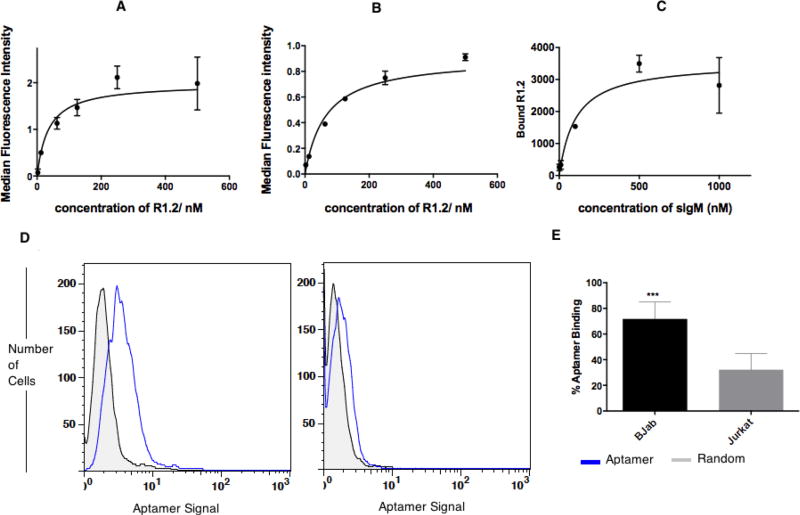
Figure 1.
Analysis of R1.2 against mIgM-positive BJAB cells. Binding affinities of R1.2 were determined by using BJAB (0.75 × 105 for 4°C and 1.5 × 105 for 37°C) cells incubated with 2.5nM to 500nM of R1.2, or random control, solutions for 45 minutes on ice (A) or 37°C (B) incubator. After washing with 3ml wash buffer, cells were reconstituted in 250 µL of wash buffer, and aptamer binding was analyzed with flow cytometry for each concentration by recording 5000 events. (C) Binding affinity of R1.2 against sIgM using nitrocellulose filter binding assay-utilizing 32P labeled R1.2 against 1µM–1nM sIgM. Serially diluted concentrations of sIgM were incubated with 32P-labelled R1.2 or random DNA for 45-minutes on ice. The bound versus unbound molecules were separated on nitrocellulose filter device by washing twice by 200 µL of wash buffer. Bound R1.2 was quantified utilizing a phosphorimager. The calculation of Bmax/2 was done as described elsewhere.12,13 (D) R1.2 shows specific binding to BJAB cells at 37°C. (E) Bar diagrams represents overall conclusion from six independent binding assays. (*** : P ≤ 0.001, obtained using student’s T-test.)