Abstract
Activation of translation initiation is a common trait of cancer cells. Formation of the heterotrimeric eukaryotic initiation factor F (eIF4F) complex is the rate-limiting step in 5′ m7GpppN cap-dependent translation. This trimeric complex includes the eIF4E cap binding protein, the eIF4G scaffolding protein, and the DEAD box RNA helicase eIF4A. eIF4A is an ATP-dependent helicase and because it is the only enzyme in the eIF4F complex, it has been shown to be a potential therapeutic target for a variety of malignancies. To this end, we have used a simple ATPase biochemical screen to survey several hundred marine and terrestrial derived natural products. Herein, we report the discovery of two natural products from marine sources, elisabatin A (1) and allolaurinterol (2), which show low µM inhibition of eIF4A ATPase activity. Enzymological analyses revealed 1 and 2 to be ATP-competitive, and cellular evaluations showed reasonable cytotoxicity against A549 (lung cancer) and MDA-MA-468 (breast cancer) cell lines. However, only compound 2 showed potent inhibition of helicase activity congruent with its ATPase inhibitory activity.
Keywords: Cancer, Translation, eIF4A, Natural products, DEAD box helicase, Inhibitor
Ongoing problems with resistance to cancer chemotherapies have led researchers to investigate the possibility of targeting nodes of regulation in cancer cells.1 A number of examples of these efforts have appeared in recent years including clinically approved proteasome inhibitors2 and compounds targeting HSP90 that are or have been in clinical trials.3 Another potential target in this genre is the DEAD-box RNA helicase associated with the eukaryotic initiation factor complex (eIF4F), called eIF4A.4 Recruitment of ribosomes to mRNA to initiate translation is a carefully regulated process requiring several protein complexes. The rate limiting step in this process is the formation of the eIF4F heterotrimeric complex that then recruits the 43S pre-initiation complex, which allows for 5′ UTR scanning of the mRNA.5 In many malignant cells, increased translational rates are observed and this is often due to increased levels of eIF4F proteins.6,7 Moreover, because the length and complexity of the 5′ UTR greatly influences eIF4A activity and many oncogenes contain complex 5′ UTRs, targeting this activity has been shown through genetic and chemical biological strategies to be a viable cancer therapeutic strategy.4,8–10 However, to date, this has not been clinically validated.
The Eukaryotic Initiation Factor 4A (eIF4A) is the founding member of the DEAD-box RNA helicases, structurally related to ATP-dependent enzymes involved in RNA processing and translation. 11–13 eIF4A forms the enzymatic core of the eIF4F translation-initiation complex, which unwinds secondary structures at the 5′ ends of mRNAs, permitting scanning for start codons by 43S ribosomal pre-initiation complexes.14 Natural products that interfere with eIF4F, such as silvestrol,15,16 pateamine A,17,18 and hippuristanol (Fig. 1),9,19 have potent anti-tumor activities and act by altering eIF4A’s interaction with RNA.8
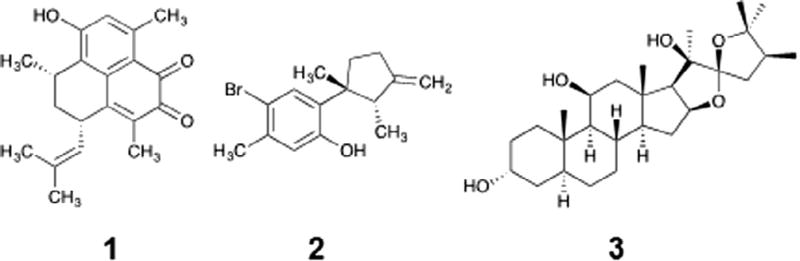
Fig. 1.
eIF4A inhibitors from an ATP hydrolysis screen against a small collection of marine derived natural products. Elisabatin A (1) was from extracts of the West Indian gorgonian octocoral Pseudopterogorgia elisabethae. Allolaurinterol (2) from dichloromethane extract of the marine red alga Laurencia obtusa. Hippuristanol (3) is a previously reported eIF4A inhibitor.
In an effort to discover lead compounds that inhibit eIF4A, a simple malachite green based assay was employed.20–22 In this assay, the release of inorganic phosphate (Pi) from ATP forms a phosphomolybdenum complex that interacts with malachite green, generating a green signal, which can be converted to the amount of inorganic phosphate, Pi. Although this assay is not as sensitive as some coupled assays23,24 or radioactive assays,4 it does not require deconvolution of the signal as coupled assays do (to account for potential inhibition of a coupling enzyme) and it is higher throughput and more environmentally friendly than radioactive assays. Evaluation of the statistical parameters of the assay revealed a robust signal with excellent statistical parameters (Z-factor >0.7; using EDTA as a positive control and DMSO as a negative control) when screening with 500 nM eIF4A and 250 µM ATP, indicating this procedure should give reliable data in a high-throughput context. As such, the malachite green assay was readily adapted to 384-well plates and used to screen 500 marine and terrestrial derived natural products. Moreover, because this is a high-throughput assay, a small panel of other DEAD box helicases was screened in parallel to look for eIF4A selective compounds. Our preliminary screen produced four hits (a 0.8% hit rate). After further investigation, we proceeded with two hits, elisabatin A (1) and allolaurinterol (2) (Fig. 1). Also, shown in Fig. 1 is the known eIF4A inhibitor hippuristanol (3). The two hits from this work, 1 and 2, were subsequently confirmed in triplicate and then advanced to an 8-point dose-response, which confirmed both compounds to be single-digit µM inhibitors of eIF4A activity (Fig. 2A and C).
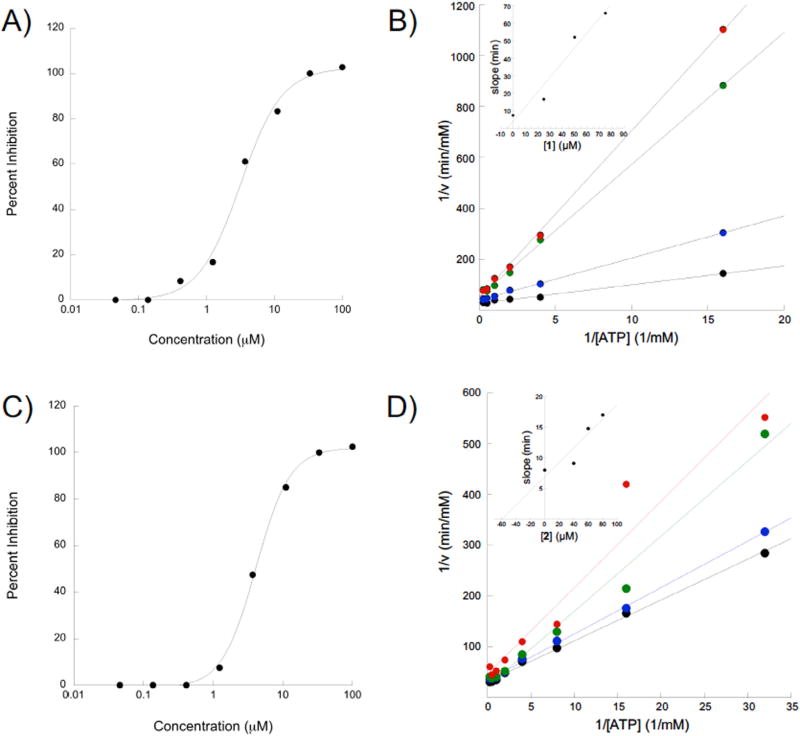
Fig. 2.
Compounds 1 and 2 inhibit eIF4A in an ATP competitive manner. A) Dose response of compound 1 and eIF4A. eIF4A ATPase activity was measured in the presence of serial dilutions of 1 and the percent inhibition was plotted as a function of [1] using a semi-log plot. B) Lineweaver-Burke analysis of 1 reveals a predominantly competitive mechanism. The inset shows a slope replot of the data as a function of inhibitor concentration. C) Dose-response for compound 2 conducted as for 1. D) Lineweaver-Burke analysis of compound 2 reveals a predominantly competitive mechanism of inhibition. The inset shows a slope replot of the data as a function of inhibitor concentration.
After confirming compounds 1 and 2 to be bona fide eIF4 A inhibitors, the mechanism of inhibition was explored using a Lineweaver-Burke analysis. To do so, four concentrations of each of the inhibitors were screened against serial dilutions of substrate (ATP) to get a series of rates. The reciprocal rates were then plotted as a function of the reciprocal substrate concentration. In this analysis, the point of intersection of the four lines informs the mechanism of inhibition. In the present case, the lines converge at the y-axis (Fig. 2B and D), which indicates an ATP-competitive mechanism of inhibition.25,26 A replot of the slopes of these lines as a function of inhibitor concentration yields a straight line that intersects the x-axis at the negative value of the Ki (Fig. 2B and D insets). The Kis for 1 and 2 are 55.04 and 3.86 µM, respectively.
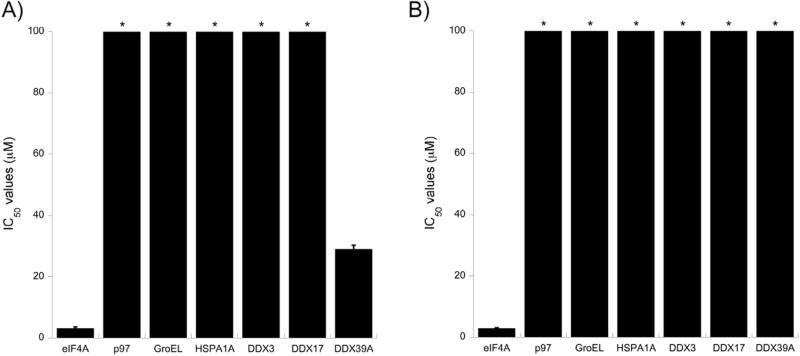
During the initial screen of the natural products collection, a panel of DEAD box helicases was evaluated, but to broaden the scope of this selectivity search, a larger panel of ATP-utilizing enzymes, including the initial DEAD box helicases, was evaluated using an 8-point serial dilution dose-response. In this analysis, standard P-loop utilizing enzymes (p97, DDX3, DDX17, and DDX39A)27 were used as well as the non-P-loop chaperones (GroEL and HSPA1A).28 As shown in Fig. 3, both compounds 1 and 2 are selective for eIF4A, with most other enzymes surveyed having IC50 values greater than 100 µM. However, there was modest inhibition of DDX39A by compound 1, although eIF4A selectivity was ~20-fold. The precise reason for compound 1 being less selective relative to DDX39A is not currently understood.
Fig. 3.
Both compound 1 and 2 show selectivity for eIF4A in biochemical assays. A), B) Compounds 1 and 2 were both measured against the panel of ATPases indicated along the x-axis. * indicates the IC50 is >100 µM.
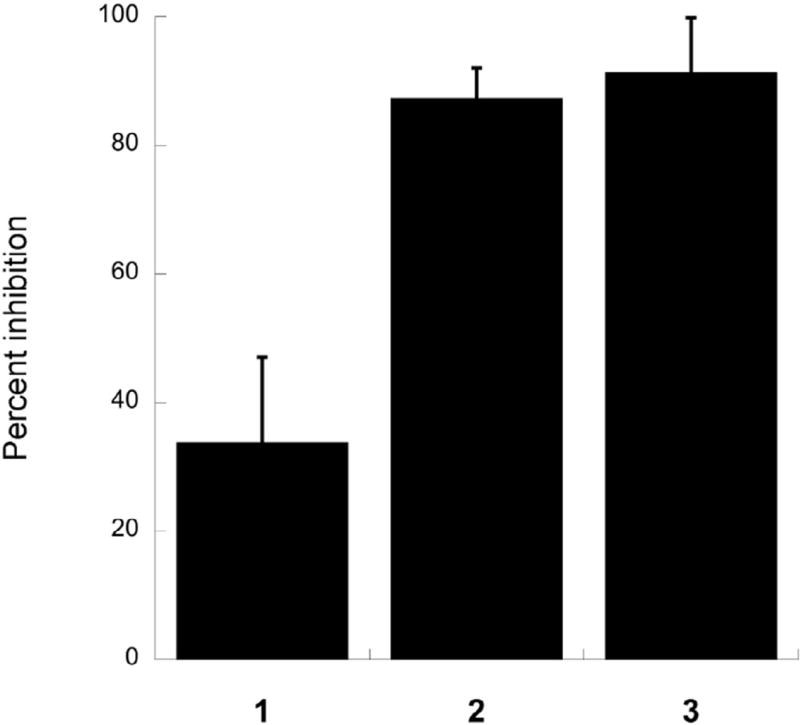
Next, the effects of compounds 1 and 2 on eIF4A mediated helicase activity were measured and compared to the known eIF4A inhibitor 3. Compound 3 is an allosteric modulator that binds to the C-terminal region of eIF4A,19 whereas 1 and 2 are ATP-competitive molecules most likely binding to the ATP-binding pocket at the interface between the N-terminal and C-terminal domains.29 It was, therefore, of interest to determine if inhibition of ATP hydrolysis blocks the RNA resolving action. The assay used for this analysis uses a fluorescently labeled strand of RNA annealed to a template next to a black-hole quencher, which blocks fluorescent signaling when both strands are annealed to the template. Upon introduction of eIF4A, eIF4B, and ATP, the fluorescent strand is released from the template, dequenching the fluorophore and providing a robust signal.30 In order to prevent rebinding to the original strand a large excess of a blank template is present. All three of the compounds were used at 100 µM. Surprisingly, even at this level, compound 1 showed only modest inhibition of eIF4A (34%) relative to a DMSO control, whereas compound 2 showed near complete inhibition of the helicase activity, at a level similar to compound 3 (Fig. 4). Presently, this difference between compounds 1 and 2 remains without an explanation.
Fig. 4.
Helicase activity assays. The ability of compounds 1 and 2 to inhibit eIF4A helicase activity were measured in a helicase assay and compared to 3. The helicase assay used measures the dequenching of a fluorophore upon release from a double strand in the Presence of eIF4A and eIF4B.
Finally, the cytotoxic activities of compounds 1 and 2 were evaluated against a lung cancer cell line (A549) and a breast cancer cell line (MDA-MA-468). A previously reported compound, elisabatin B, which was isolated from the same organism as 1, had been reported to have cytotoxic activity against a variety of cancers in the NCI-60 panel,31 but only modest activity was reported for compound 1 against the NCI-60 panel.33 Compound 2 has been previously shown to have antibiotic activity, but had not been reported to have cytotoxic activity against cancer cell lines.32 However, given the current discovery that compounds 1 and 2 target eIF4A, it was expected that they would have cytotoxic activity. To this end, an MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay was used to determine the LD50 of both compounds using an 8-point dose response (Fig. 5). Both compounds showed modest toxicity against A549 (40.2 µM and 41.4 µM) and MDA-MA-468 (35.2 µM and 31.7 µM) cell lines. These values are approximately 10-fold higher than the IC50 values. A number of factors may contribute to this difference including limited cellular penetrance or metabolism. Although these studies do not indicate eIF4A to be the only cellular target of 1 and 2, it is likely a mode of cellular action. Improvement of the pharmacology and potency of 1 and 2 will be reported at a later date.
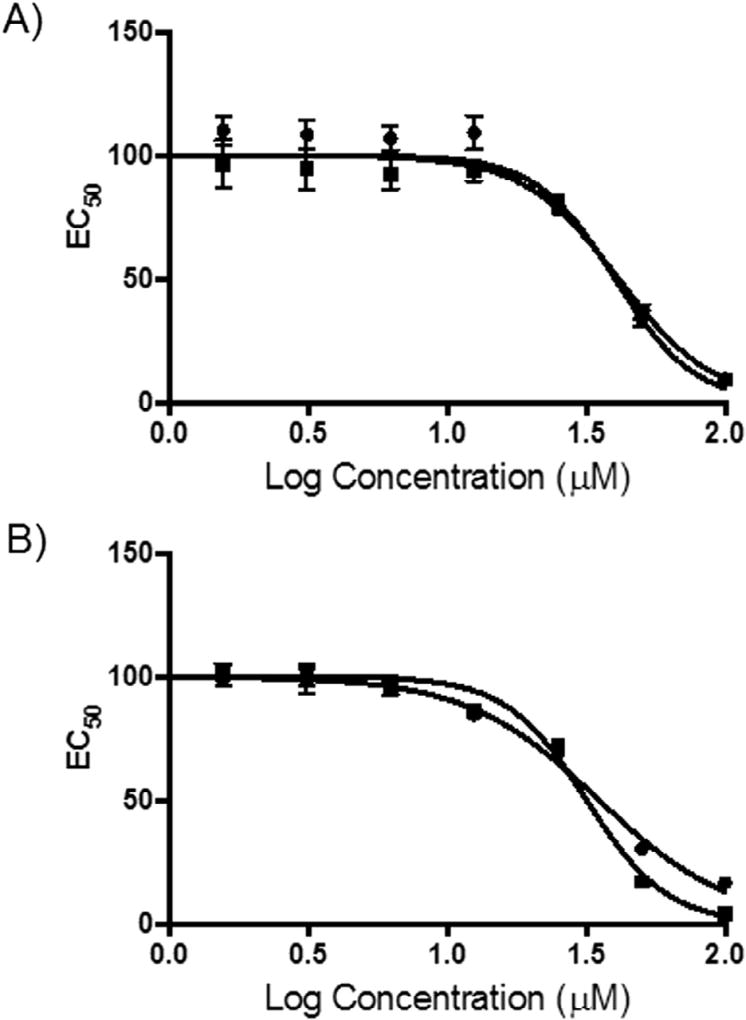
Fig. 5.
Cytotoxicity of compounds 1 and 2. A) An 8-point dose-response against A549 lung cancer cells. Cell viability was measured using an MTT assay and percent survival was plotted as a function of compound concentration on semi-log axes. The LD50 values are 40.2 µM and 41.4 µM for 1 and 2, respectively. B) Same as A) except MDA-MA-468 breast cancer cells were used. The LD50 values are 35.2 µM and 31.7 µM for 1 and 2, respectively. Circles indicate compound 1 and squares are compound 2.
Supplementary Material
Acknowledgments
This work was supported by funds from the University of Arizona (E.C.), the National Institutes of Health (NIH) Training Grant T32 GM008804 (A.J.A.), the National Institute of Environmental Health Sciences Training Grant T32 ES007091 (J.T.) and National Institute of Health (NIH) Training Grant T32 HL007249 (M.K.).
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.bmcl.2017.07.045.
References
- 1.Nagarajan A, Malvi P, Wajapeyee N. Trends Cancer. 2016;2:365–377. doi: 10.1016/j.trecan.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moreau P, Richardson PG, Cavo M, et al. Blood. 2012;120:947–959. doi: 10.1182/blood-2012-04-403733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garg G, Khandelwal A, Blagg BS. Adv Cancer Res. 2016;129:51–88. doi: 10.1016/bs.acr.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cencic R, Galicia-Vazquez G, Pelletier J. Methods Enzymol. 2012;511:437–461. doi: 10.1016/B978-0-12-396546-2.00020-6. [DOI] [PubMed] [Google Scholar]
- 5.Sonenberg N, Hinnebusch AG. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silvera D, Formenti SC, Schneider RJ. Nat Rev Cancer. 2010;10:254–266. doi: 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- 7.Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, Topisirovic I. Nat Rev Drug Discovery. 2015;14:261–278. doi: 10.1038/nrd4505. [DOI] [PubMed] [Google Scholar]
- 8.Chu J, Pelletier J. Biochim Biophys Acta. 2015;1849:781–791. doi: 10.1016/j.bbagrm.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Lindqvist L, Pelletier J. Future Med Chem. 2009;1:1709–1722. doi: 10.4155/fmc.09.122. [DOI] [PubMed] [Google Scholar]
- 10.Schatz JH, Oricchio E, Wolfe AL, et al. J Exp Med. 2011;208:1799–1807. doi: 10.1084/jem.20110846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andreou AZ, Klostermeier D. RNA Biol. 2013;10:19–32. doi: 10.4161/rna.21966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linder P, Fuller-Pace FV. Biochim Biophys Acta. 2013;1829:750–755. doi: 10.1016/j.bbagrm.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 13.Shadrick WR, Ndjomou J, Kolli R, Mukherjee S, Hanson AM, Frick DN. J Biomol Screen. 2013;18:761–781. doi: 10.1177/1087057113482586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marintchev A. Biochim Biophys Acta. 2013;1829:799–809. doi: 10.1016/j.bbagrm.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bordeleau ME, Robert F, Gerard B, et al. J Clin Invest. 2008;118:2651–2660. doi: 10.1172/JCI34753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cencic R, Carrier M, Galicia-Vazquez G, et al. PLoS One. 2009;4:e5223. doi: 10.1371/journal.pone.0005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bordeleau ME, Matthews J, Wojnar JM, et al. Proc Natl Acad Sci USA. 2005;102:10460–10465. doi: 10.1073/pnas.0504249102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bordeleau ME, Cencic R, Lindqvist L, et al. Chem Biol. 2006;13:1287–1295. doi: 10.1016/j.chembiol.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 19.Bordeleau ME, Mori A, Oberer M, et al. Nat Chem Biol. 2006;2:213–220. doi: 10.1038/nchembio776. [DOI] [PubMed] [Google Scholar]
- 20.Kang MJ, Wu T, Wijeratne EM, et al. ChemBioChem. 2014;15:2125–2131. doi: 10.1002/cbic.201402258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henkel RD, VandeBerg JL, Walsh RA. Anal Biochem. 1988;169:312–318. doi: 10.1016/0003-2697(88)90290-4. [DOI] [PubMed] [Google Scholar]
- 22.Kagami O, Kamiya R. Methods Cell Biol. 1995;47:147–150. doi: 10.1016/s0091-679x(08)60803-1. [DOI] [PubMed] [Google Scholar]
- 23.Jenkins WT. Anal Biochem. 1991;194:136–139. doi: 10.1016/0003-2697(91)90160-u. [DOI] [PubMed] [Google Scholar]
- 24.Penefsky HS, Pullman ME, Datta A, Racker E. J Biol Chem. 1960;235:3330–3336. [PubMed] [Google Scholar]
- 25.Lineweaver H, Burk D. J Am Chem Soc. 1934;56:658–666. [Google Scholar]
- 26.Dixon M. Biochem J. 1972;129:197–202. doi: 10.1042/bj1290197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saraste M, Sibbald PR, Wittinghofer A. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 28.Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Annu Rev Biochem. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 29.Schutz P, Bumann M, Oberholzer AE, et al. Proc Natl Acad Sci USA. 2008;105:9564–9569. doi: 10.1073/pnas.0800418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ozes AR, Feoktistova K, Avanzino BC, Baldwin EP, Fraser CS. Nat Protoc. 2014;9:1645–1661. doi: 10.1038/nprot.2014.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodriguez AD, Gonzalez E, Huang SD. J Org Chem. 1998;63:7083–7091. doi: 10.1021/jo981385v. [DOI] [PubMed] [Google Scholar]
- 32.Konig GM, Wright AD. Planta Med. 1997;63:186–187. doi: 10.1055/s-2006-957643. [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez AD, Ramirez C, Rodriguez II. J Nat Prod. 1999;62:997–999. doi: 10.1021/np990090p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.