Abstract
Ewing sarcoma (ES) is the most common malignant bone tumor in children and young adults. It is characterized by chromosomal translocations fusing the EWS gene with an ETS oncogene, most frequently FLI1. In the present study, the authors aimed to investigate the function of EWS-FLI1 in autophagy in ES cells, and identified that EWS-FLI1 positively regulates autophagy in ES cells. ATG4B expression was observed markedly upregulated by EWS-FLI1 overexpression, and silencing of ATG4B dramatically inhibits autophagy in ES cells. Furthermore, apoptosis was inhibited in ATG4B overexpressed ES cells, and ATG4B-potentiated autophagy is required for ES cells survival. Taken together, the authors demonstrated the role of EWS-FLI1 and ATG4B in autophagy in ES cells, and suggested EWS-FLI1 and ATG4B as potential therapeutic targets for ES.
Keywords: EWS-FLI1, ATG4B, Ewing sarcoma, autophagy
Introduction
Ewing sarcoma (ES) is a highly malignant bone and soft tissue tumor that most often occur in children, teenagers and young adults with a peak incidence between the ages of 14 and 20 years (1,2). While the histogenesis of ES remains enigmatic, 85% of cases are defined by the t(11;22)(q24;q12) chromosomal translocation, resulting in a pathognomonic chimeric fusion gene which encodes the EWS-FLI1 protein (3). EWS-FLI1 induces massive deregulation of protein expression by either transcriptionally inducing or repressing specific target genes, many of which are involved in the oncogenic process, including cell proliferation (4), transformation (5) and in vivo tumor growth (6), but the function of EWS-FLI1 in autophagy still remains unknown.
Macroautophagy (hereafter referred to as autophagy) has proven to be an evolutionarily conserved catabolic pathway within all eukaryotic cells that plays a critical function for cellular homeostasis to remove senescent or damaged proteins and organelles through lysosome-mediated degradation (7,8). In addition to removing dysfunctional proteins and organelles, autophagy could also provide amino acids, monosaccharides, nucleic acids and lipids during times of nutrient deprivation (9,10). Autophagy can be stimulated by various stressors, such as starvation, oxidative stress and pharmaceutical compound treatment (11). Besides maintaining normal cellular homeostasis, autophagy was also illustrated to play crucial roles in infectious, inflammatory, neurodegenerative and metabolic diseases (12–15). Moreover, autophagy was identified to have dual roles in tumors (16). In some cases, autophagy was found to serve a tumor suppressor function (17,18). Paradoxically, however, accumulating evidence strongly suggests that autophagy promotes tumor growth and resistance to chemotherapy in established tumors (19,20). These two distinct functions may be mainly due to different genes induced by autophagy in different tumors and conditions (21).
At least 36 autophagy-related (ATG) genes are primarily involved in the progression of autophagy from phagophore initiation to autophagosome formation in mammalian cells (22). Among these, ATG7 and ATG3 conjugate mammalian light chain 3 (LC3) homologues to phosphatidyl ethanolamine (PE), and ATG7 and ATG10 conjugate ATG12 to ATG5 (23,24). The cysteine protease ATG4 contributes to this chain of events by cleaving the LC3 C-terminal domain to generate LC3-I (25). Consequently, LC3-I is converted by ATG7 and ATG3 to LC3-II, which is essential for phagophore and autophagosome formation (22,26). And the LC3-II on the cytoplasmic location of autolysosome is recycled following delipidation of PE, which is also performed by ATG4 (27). There are four different ATG4 paralogs expressed in mammals (ATG4A, ATG4B, ATG4C and ATG4D) with ATG4B being crucial for the autophagic process and having the broadest substrate spectrum for different LC3 forms and homologs (28,29).
In the present study, the authors investigated the function of EWS-FLI1 in autophagy in ES cells. It was identified that autophagy was promoted by overexpressing EWS-FLI1 in ES cells, as evidenced by the decreases in the amount of p62 (SQSTM1) and increases in the amount of LC3B-II, two important markers of autophagy, as well as the increased LC3 puncta in cells. Furthermore, ATG4B was significantly upregulated by EWS-FLI1 overexpression, and ATG4B gene may be a transcript target of EWS-FLI1. To the best of the authors' knowledge, the present study is the first to have examined the function of ATG4B in ES cells. ATG4B was shown to greatly modulate the autophagy process and ATG4B-potentiated autophagy is required for ES cells survival. In conclusion, the authors revealed the effect of EWS-FLI1 and ATG4B on autophagy in ES cells in the present study, and suggested EWS-FLI1 and ATG4B as potential therapeutic targets for ES.
Materials and methods
Cell culture
NIH3T3, A673 and TC71 cells were obtained from American Type Culture Collection (Manassas, VA, USA). All the cells were cultured in RPMI-1640 (Gibco, Life Technologies, Shanghai, China) containing 10% fetal calf serum (Gibco, Life Technologies, Mulgrave, VIC, Australia) for normal condition. Cells were cultured in serum-free medium for starvation for indicated time to induce autophagy.
Lentivirus and plasmid
Lentiviruses containing the following were constructed and obtained from MDL Biotechnology (Beijing, China): An empty plasmid or expression plasmid of EWS-FLI1, control small interfering (si)RNA and EWS-FLI1 siRNAs, control plasmid and ATG4B expression plasmid, short hairpin (sh)RNA-control plasmid and shRNA-ATG4B plasmid. The lentivirus particle was used to infect NIH3T3 for 3 days at 50 MOI with the presence of polybrene, puromycin selection was performed to establish the overexpression cell lines. Lipofectamine 2000 transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used to transfect plasmids and siRNAs into ES cells.
Western blot analysis
The cells were harvested by using radioimmunoprecipitation assay buffer (MDL Biotechnology), and the protein concentration was measured using Bio-Rad quantification assay (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Proteins were separated using 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, USA). The membranes were blocked with 3 g non-fat dry milk in TBST (50 mM Tris-HCl, 150 mM NaCl, 0.1% Tween-20, pH 7.4) for 2 h. Primary antibodies including anti-p62 (1:500, #sc-101542), anti-LC3 (1:500, #sc-398822), anti-FLI1 (1:500, #sc-22808), anti-cyt c (1:500, #sc-13561), anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (1:1,000, #sc-32233) were bought from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA) and anti-ATG4B (1:500, #13507), anti-PUMA (1:500, #4976) were obtained from Cell Signaling Technology (Beverly, MA, USA). The antibodies were added and incubated overnight at 4°C. After washing, the membranes was incubated with anti-mouse secondary antibody (1:2,000, #sc2055) or anti-rabbit secondary antibody (1:2,000, #sc2054) (both from Santa Cruz Biotechnology, Inc.) at room temperature for 1 h. The target protein was visualized by enhanced chemiluminescence (Thermo Fisher Scientific, Inc.). The results from western blot analysis were quantitatively analyzed by optical density using ImageJ software.
RNA Isolation, reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis and CHIP assay
Total RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. A LightCycler (ABI PRISM 7000; Applied Biosciences; Thermo Fisher Scientific, Inc.) and a SYBR RT-PCR kit (Takara Biotechnology Co., Ltd., Dalian, China) were used for RT-qPCR analysis. GAPDH was used as the internal control, thermocycling conditions were 1 cycle (95°C, 5 min) and 40 cycles (95°C, 15 sec; 57°C, 30 sec; 72°C, 30 sec) and the 2−ΔΔCt method (30) was used to evaluate the relative quantities of each amplified product in the samples. CHIP assay was performed as previously reported and the primer sequences for qPCR and CHIP-qPCR were used as described (31).
Dual-luciferase reporter gene assay
ATG4B promoter region was cloned into pGL3-based vectors to construct the luciferase reporter plasmid. The phRL-TK plasmid was used as an internal control. The luciferase activities were measured on a SpectraMax M5 reader (Molecular Devices LLC, Sunnyvale, CA, USA) using the Dual Luciferase Reporter assay system (Promega Corp., Madison, WI, USA).
Confocal microscopic analysis
The experiment was performed as described (31). All slides were observed under a confocal laser microscopy (LSM780; Carl Zeiss AG, Oberkochen, Germany).
Cell viability assay and detection of cell apoptosis
Cell viability was examined by CCK8 kit and cell apoptosis percent was detected by flow cytometry as described (32). The caspase-3 activity of cell extracts (~106 cells) was determined using the synthetic substrate, acetyl-Asp-Glu-Val-Asp-7-amino-4-methyl coumarin (DEVD-AMC; Peptron, Inc., Daejeon, Korea) according to the manufacturer's instructions. The AMC fluorescence released by active caspase-3 was measured at an excitation wavelength of 360 nm and an emission wavelength of 460 nm.
Statistical analysis
All data are presented as mean ± standard deviation of three independent experiments. Statistical significance was determined with the two-tailed Student's t-test to compare two groups. One-way analysis of variance (ANOVA) was performed to compare three or more groups. If the ANOVA analysis was significant, the Newman-Keuls test was applied for comparison between each two groups. P<0.05 was considered to indicate a statistically significant difference.
Results
EWS-FLI1 promotes autophagy in ES cells
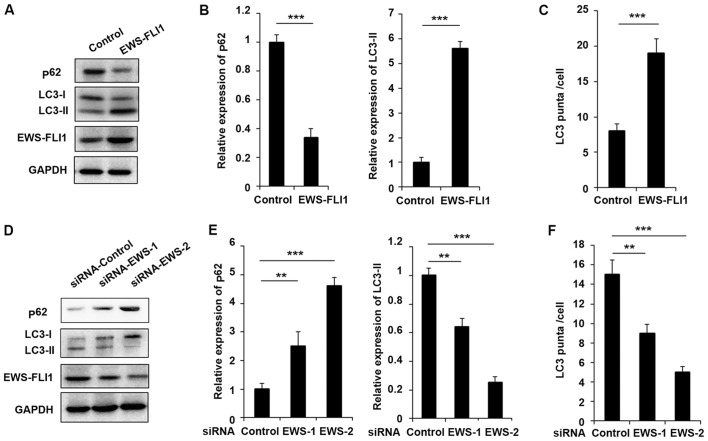
In order to investigate the function of EWS-FLI1 in autophagy in ES, the authors used in vitro cultured NIH3T3 cells and ES tumor-derived cell lines to perform the study. First, the lentivirus containing EWS-FLI1 expressing plasmid was used to construct NIH3T3-EWS-FLI1 stable overexpression cell lines. As presented in Fig. 1A, EWS-FLI1 expression was significantly increased after EWS-FLI1-lentivirus infection compared to control-lentivirus infection. Most important, the expression of p62 was decreased and the level of LC3-II was increased in EWS-FLI1 overexpressed NIH3T3 cells (Fig. 1A and B). Furthermore, the authors used EWS-FLI1 specific siRNA to knockdown the expression of EWS-FLI1 in A673 cells. As presented in Fig. 1D, the effect of the two siRNAs was examined and EWS-FLI1 expression was much weaker in siRNA-2 treated A673 cells compared to cells transfected with siRNA-1 (Fig. 1D and E). Interestingly, the expression of p62 was significantly enhanced and LC3-II expression was markedly decreased in both siRNAs treated cell, consistent with the knockdown efficiency (Fig. 1D and E). The effect of EWS-FLI1 on autophagy was also verified by immunofluorescence to show the formation of autophagosomes labeled the by anti-LC3 antibody, and the observed stable overexpression of EWS-FLI1 in NIH3T3 greatly increased the formation of autophagosomes, accordant results were obtained in EWS-FLI1 silencing A673 cells (Fig. 1C and F).
Figure 1.
EWS-FLI1 promotes autophagy in ES cells. (A) Western blot analysis of p62, LC3-I, LC3-II, EWS-FLI1 and GAPDH in NIH3T3 cells stable over-expressing control plasmid or EWS-FLI1 expression plasmid following starvation for 2 h. (B) Band intensity of p62 and LC3-II was quantified and normalized to the GAPDH in (A). (C) After starvation for 2 h, LC3 puncta in NIH3T3 cells stable overexpressing control plasmid or EWS-FLI1 expression plasmid was detected by confocal laser microscopy and quantified using the ImageJ program, 30 to 60 cells chosen in random were counted. (D) Western blot analysis of p62, LC3-I, LC3-II, EWS-FLI1 and GAPDH in control-siRNA or EWS-FLI1-siRNAs treated A673 cells after starvation for 2 h. (E) Band intensity of p62 and LC3-II was quantified and normalized to the GAPDH in (D). (F) After starvation for 2 h, LC3 puncta in control-siRNA or EWS-FLI1-siRNAs treated A673 cells were detected by confocal laser microscopy and quantified using the ImageJ program, 30 to 60 cells chosen in random were counted. Data are representative of three independent experiments (mean ± standard deviation). **P<0.01 and ***P<0.001 as indicated. ES cells, Ewing sarcoma cells; siRNA, small interfering RNA; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
ATG4B is a transcript target of EWS-FLI1
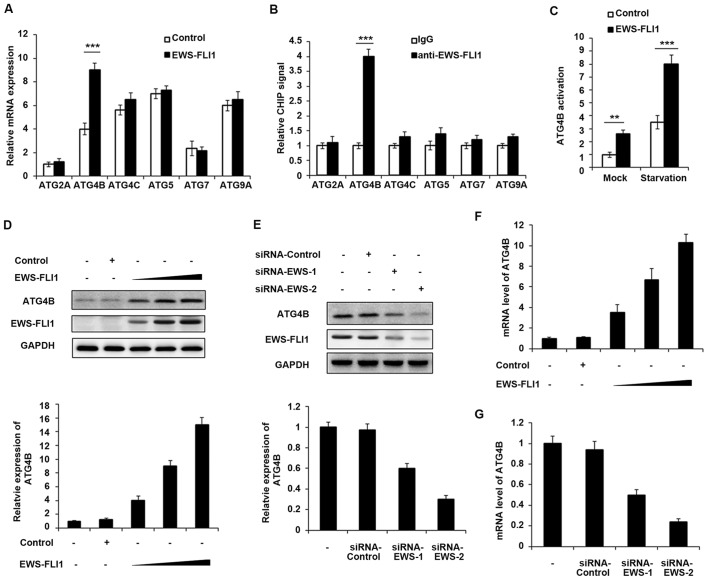
EWS-FLI1 induces massive deregulation of protein expression, therefore the authors aimed to examine whether EWS-FLI1 could regulate the expression of ATG proteins. The expression of various ATG proteins was investigated including ATG2A, ATG4B, ATG4C, ATG5, ATG7 and ATG9A, and only ATG4B expression was significantly upregulated by EWS-FLI1 overexpression (Fig. 2A). Consistently, the CHIP-qPCR result demonstrated that EWS-FLI1 has the ability to interact with ATG4B promoter but not others (Fig. 2B), which meant that ATG4B gene may be a transcript target of EWS-FLI1. In addition, ATG4B activation was significantly increased in EWS-FLI1 stable overexpressed NIH3T3 cells, especially in starvation conditions (Fig. 2C). Moreover, the authors infected NIH3T3 cells with increasing titer of EWS-FLI1-lentivirus, and the expression of ATG4B was upregulated in a dose-dependent manner in both protein and mRNA levels (Fig. 2D and F). Consistent with the overexpression data, knockdown the expression of EWS-FLI1 also decreased the mRNA and protein levels of ATG4B (Fig. 2E and G).
Figure 2.
ATG4B is a transcript target of EWS-FLI1. (A) RT-qPCR analysis of mRNA expression of ATG2A, ATG4B, ATG4C, ATG5, ATG7 and ATG9A in NIH3T3 cells stable overexpressing control plasmid or EWS-FLI1 expression plasmid after starvation for 2 h. (B) CHIP assay and RT-qPCR analysis were performed with fragmented chromatin from NIH3T3 cells stable overexpressing control plasmid or EWS-FLI1 expression plasmid after starvation for 2 h. (C) Dual-luciferase reporter gene assay was performed to investigate the activation of ATG4B promoter in NIH3T3 cells stable overexpressing control plasmid or EWS-FLI1 expression plasmid in normal or starvation condition for 2 h. (D) NIH3T3 cells were infected with increasing titer of EWS-FLI1-lentivirus, the protein levels of ATG4B and EWS-FLI1 were examined by western blotting, and band intensity of ATG4B was quantified and normalized to the GAPDH. (E) A673 cells were treated with control-siRNA or EWS-FLI1 siRNAs, the protein levels of ATG4B and EWS-FLI1 were examined by western blot, and band intensity of ATG4B was quantified and normalized to the GAPDH. (F and G) RT-qPCR analysis of mRNA level of ATG4B in (D and E). Data are representative of three independent experiments (mean ± standard deviation). **P<0.01 and ***P<0.001 vs. control. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; siRNA, small interfering RNA; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Overexpression of ATG4B promotes autophagy in ES cells
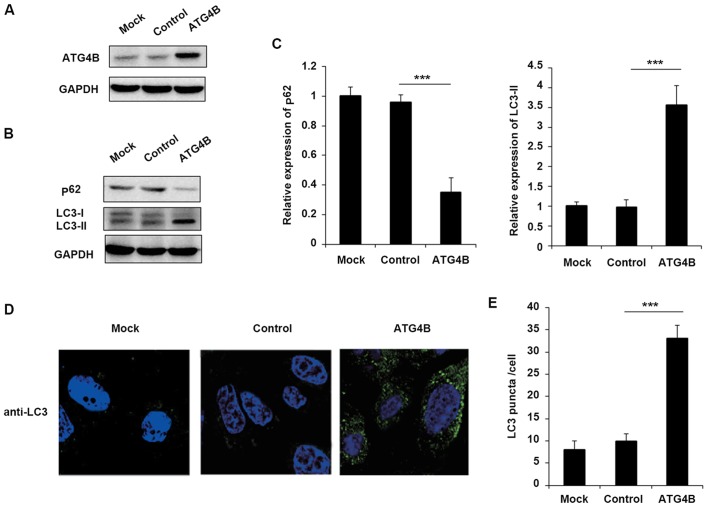
Although ATG4B has been reported that serve essential roles in autophagy in numerous cells, the function of ATG4B in ES cells still remains unknown. To confirm that EWS-FLI1 affected autophagy through ATG4B, the authors further examined the effect of ATG4B on autophagy in ES cells. Overexpression of ATG4B in TC71 cells was confirmed as presented in Fig. 3A. Consistent with the results of EWS-FLI1 overexpression, ATG4B overexpression also decreased the level of p62 and enhanced the expression of LC3-II (Fig. 3B and C), as well as the increased formation of autophagosomes (Fig. 3D and E).
Figure 3.
Overexpression of ATG4B promotes autophagy in ES cells. (A) TC71 cells were transfected with ATG4B expression plasmid or control plasmid, and overexpression of ATG4B was examined by western blotting. (B) Western blot analysis of p62, LC3-I, LC3-II and GAPDH in ATG4B overexpressed TC71 cells following starvation for 2 h. (C) Band intensity of p62 and LC3-II were quantified and normalized to the GAPDH in (B). (D) TC71 cells transfected with ATG4B expression plasmid or control plasmid were inspected under confocal laser microscopy to detect LC3 puncta by immunofluorescence after starvation for 2 h. Magnification, ×400. (E) LC3 puncta number per cell in (D) was quantified using the ImageJ program, and 30–60 cells chosen in random were counted. Data are representative of three independent experiments (mean ± standard deviation). ***P<0.001 as indicated. ES cells, Ewing sarcoma cells; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Silencing of ATG4B inhibits autophagy in ES cells
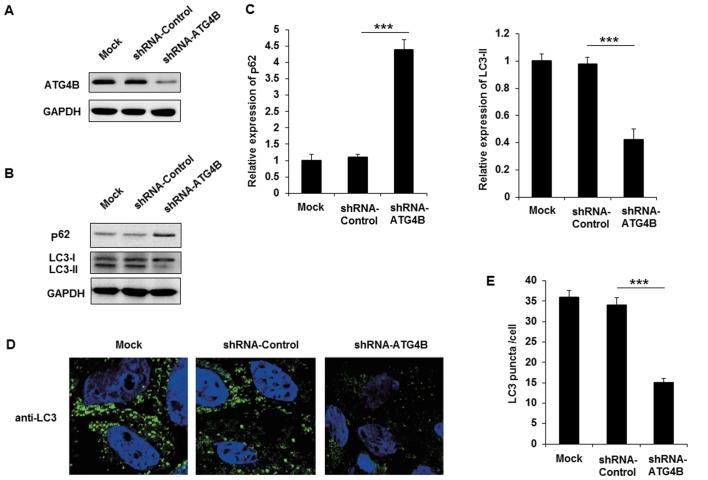
The authors purchased shRNA plasmid of ATG4B to knockdown the expression of ATG4B in A673 cells, and the effect of ATG4B specific shRNA was examined (Fig. 4A). Following transfection with shRNA-ATG4B, the expression of p62 was markedly increased and LC3-II level was decreased (Fig. 4B and C). Similar to the EWS-FLI1 silencing results, knockdown the expression of ATG4B also decreased autophagosomes formation in A673 cells. Together with the data in ATG4B overexpressed TC71 cells, these results indicated that ATG4B indeed promoted the autophagy process in ES cells.
Figure 4.
Silencing of ATG4B inhibits autophagy in ES cells. (A) A673 cells were transfected with shRNA-ATG4B plasmid or shRNA-control plasmid, and the expression of ATG4B was examined by western blotting. (B) Western blot analysis of p62, LC3-I, LC3-II and GAPDH in ATG4B silenced A673 cells following starvation for 4 h. (C) Band intensity of p62 and LC3-II were quantified and normalized to the GAPDH in (B). (D) A673 cells transfected with shRNA-ATG4B plasmid or shRNA-control plasmid were inspected under confocal laser microscopy to detect LC3 puncta by immunofluorescence after starvation for 4 h. Magnification, x400. (E) LC3 puncta number per cell in (D) was quantified using the ImageJ program, and 30-60 cells chosen in random were counted. Data are representative of three independent experiments (mean ± standard deviation). ***P<0.001 as indicated. ES cells, Ewing sarcoma cells; shRNA, short hairpin RNA; GAPDH, glyceraldehyde 3-phosphate dehydrogenase
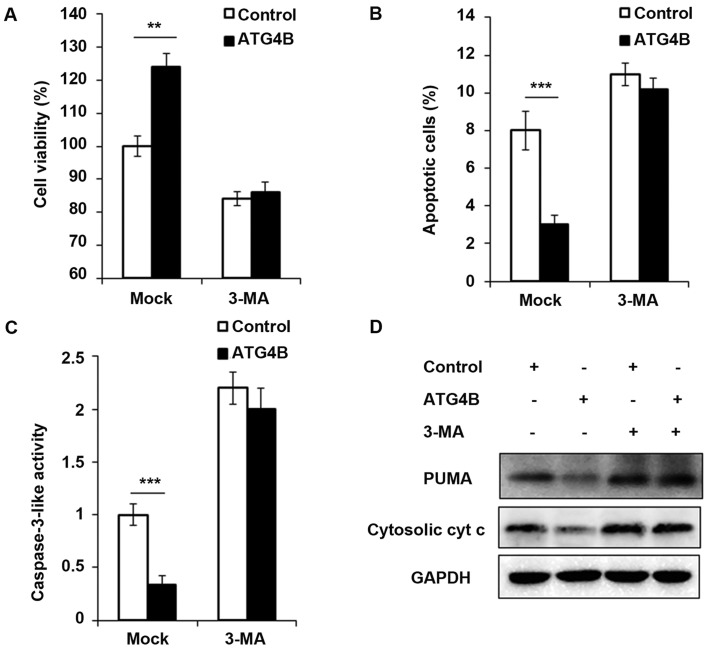
Cell viability is increased and apoptosis is attenuated in ATG4B overexpressed ES cells
In addition, the authors investigated the contribution of ATG4B-mediated autophagy to tumor cell viability and apoptosis. As presented in Fig. 5A, ATG4B overexpressed TC71 cells with high level of autophagy grew significantly faster than TC71 cells transfected with control plasmid. However, the blockage of autophagy by 3-methyladenine (3-MA), an important agent to block autophagy, retarded the cell viability of ATG4B overexpressed cells. Apoptosis is suppressed by autophagy in various cancer cells (33,34). Recently, it has been reported that ATG4B protease and autophagy serve crucial roles in protecting epithelial cells against bleomycin-induced pulmonary fibrosis and apoptosis (35). Therefore, the authors investigated the effect of ATG4B-induced autophagy on apoptosis in ES cells. As presented in Fig. 5B and C, the number of apoptotic cells and caspase-3 activity was greatly decreased in ATG4B-overexpressed cells, treatment of 3-MA increased the apoptosis process as previous study reported (36), however after 3-MA treatment, there was no significant difference of apoptosis between ATG4B overexpressed cells and control group (Fig. 5B and C). Furthermore, the authors examined the expression of PUMA and cytosolic cytochrome c, and both the protein levels of PUMA and cytosolic cytochrome c was found decreased in ATG4B overexpressed cells without 3-MA challenge, nevertheless, the downregulated expression of PUMA and cytosolic cytochrome c modulated by ATG4B overexpression was all observed to be reversed following 3-MA treatment.
Figure 5.
Cell viability is increased and apoptosis is attenuated in ATG4B overexpressed ES cells. (A) Following starvation for 48 h, cell viability of TC71 cells transfected with control plasmid or ATG4B expression plasmid was measured by CCK8 assay in with or without 3-MA to block autophagy for 24 h. (B and C) After starvation for 48 h, the percentage of apoptotic cell and caspase-3 activity of TC71 cells was measured by flow cytometry or fluorometric assay with or without 3-MA to block autophagy for 24 h. (D) Western blot analysis of PUMA, cytosolic cytochrome c and GAPDH in TC71 cells transfected with control plasmid or ATG4B expression plasmid after starvation for 2 h. Data are representative of three independent experiments (mean ± standard deviation). **P<0.01 and ***P<0.001 as indicated. 3-MA, 3-methyladenine; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
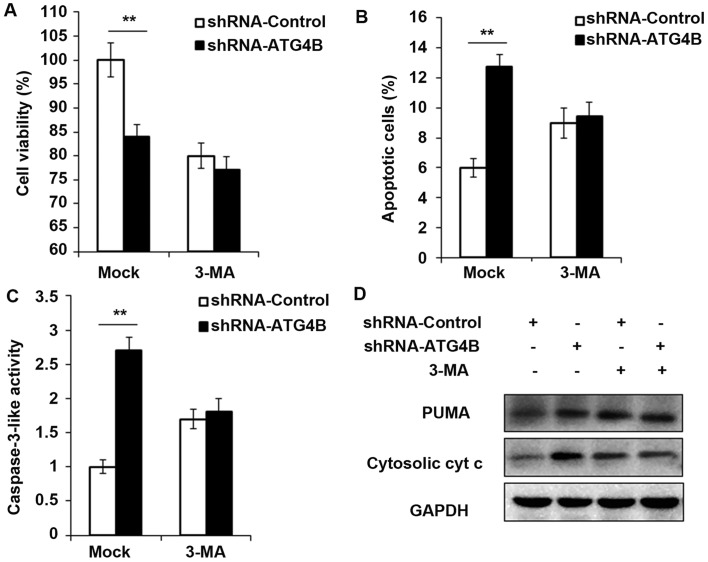
Cell viability is decreased and apoptosis is potentiated in ATG4B silenced ES cells
To ensure the results in Fig. 5, shRNA-ATG4B was used to repeat the experiments, and consistent with the ATG4B overexpression results, knockdown the expression of ATG4B indeed decreased the cell viability and increased the apoptotic cell number (Fig. 6A and B), as well as the activity of caspase-3 and expression of PUMA and cytosolic cytochrome c (Fig. 6C and D). But all these processes were found reversed after autophagy was blocked by 3-MA.
Figure 6.
Cell viability is decreased and apoptosis is potentiated in ATG4B silenced ES cells. (A) Following starvation for 48 h, cell viability of A673 cells transfected with shRNA-ATG4B plasmid or shRNA-control plasmid was measured by CCK8 assay in with or without 3-MA to block autophagy for 24 h. (B and C) After starvation for 48 h, the percentage of apoptotic cell and caspase-3 activity of A673 cells transfected with shRNA-ATG4B plasmid or shRNA-control plasmid was measured by flow cytometry or fluorometric assay with or without 3-MA to block autophagy for 24 h. (D) Western blot analysis of PUMA, cytosolic cytochrome c and GAPDH in A673 cells transfected with shRNA-ATG4B plasmid or shRNA-control plasmid after starvation for 2 h. Data are representative of three independent experiments (mean ± standard deviation). **P<0.01 as indicated. ES cells, Ewing sarcoma cells; shRNA, short hairpin RNA; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Discussion
In the present study, the authors investigated the function of EWS-FLI1 in autophagy in ES. The present study may extend our understanding of the pathogenesis of ES, and provided a new approach for the related drug designs.
Described for the first time in 1921 by Ewing (37), ES primarily arises at the pelvic bones, the diaphysis of the lower extremities' long bones and the chest wall's bones. The overall survival of ES patients remains poor: 25% of patients with localized tumor and 75% of patients with metastasis do not have durable therapeutic responses (38). ES is characterized by unique chromosomal translocations and EWS-FLI1 is the most commonly fusion protein observed in ES (39). Transfection of EWS-FLI1 into NIH3T3 cells was reported to lead to the phenotypic characteristics of ES (40). In the present study, to extend our understanding of EWS-FLI1 in autophagy, a lentivirus containing a EWS-FLI1 expression plasmid was used to build a EWS-FLI1 stable expression NIH3T3 cell line. Overexpression of EWS-FLI1 was identified in NIH3T3 cells leading to the attenuated p62 expression and enhanced LC3-II expression, as well as the autophagosomes number is significantly increased. Silencing EWS-FLI1 resulted in the consistent data. Based on these results, the authors supposed that EWS-FLI1 could regulate the autophagy process in ES.
Autophagy facilitates the clearance of long-lived proteins, aggregates and damaged organelles, and dysregulation of autophagy is highly associated with cancer, particularly in tumorigenesis and chemotherapy resistance (39,41). The cysteine protease ATG4B serves a role in key steps of the autophagy process as cleaving proLC3 isoforms to form LC3-I for subsequent lipidation to form LC3-II and autophagosome membrane insertion. A previous study demonstrated that ATG4B was essential for starvation-induced autophagy, and inhibition of ATG4B activity by anti-autophagy compound NSC185058 greatly attenuated osteosarcoma tumorigenesis (42). In addition, ATG4B was reported to promote colorectal cancer growth and suggested as a potential biomarker and therapeutic target in CML stem/progenitor cells (43,44). Nevertheless, the study of ATG4B in ES remains largely unknown. In the present study, the authors found that EWS-FLI1 induced the expression of ATG4B and EWS-FLI1 was observed to interact with the ATG4B promoter, which suggested that ATG4B as a transcriptional target of EWS-FLI1. In addition, overexpression of ATG4B significantly promoted the autophagy in ES cells, consistently, silencing of ATG4B markedly suppressed the progression of autophagy. These results supposed that the overexpression of EWS-FLI1 may also lead to the increased expression of ATG4B in ES tissues, however, we still need to make further studies to confirm the exact conclusion.
The profound relationship between autophagy and apoptosis has been widely reported. Autophagy was found to promote apoptosis in response to specific stimuli (45), but accumulated evidences showed that autophagy could protect against apoptosis (46,47). In the present study, the authors also examined the function of ATG4B in cell viability. Consistent with the previous results in other cell types (43,44), overexpression of ATG4B was found to promote cell growth in ES cells, but blockade of ATG4B-dependent autophagic flux was insufficient to accelerate cell growth in ES cells. Furthermore, the role of ATG4B on apoptosis was investigated and ATG4B dramatically suppressed the apoptosis process in ES cells. In addition, it was found that ATG4B-dependent autophagic flux is also necessary for the inhibition of apoptosis. Moreover, the authors examined the level of PUMA and cytosolic cytochrome c, two important pro-apoptotic proteins, and both the expression of PUMA and cytosolic cytochrome c was significantly decreased in ATG4B overexpressed ES cells, but the processes were found reversed after autophagy was blocked by 3-MA. Taken together, these findings suggested that ATG4B-dependent autophagy plays great roles in regulating cell viability and cell apoptosis in ES cells.
In conclusion, in the present study, the authors revealed the promoting effect of EWS-FLI1 and ATG4B on autophagy in ES cells, and these findings may provide new venues for anti-autophagy drugs and new targets for the treatment of ES.
Acknowledgments
This study was supported by grants from the Natural Science Foundation of Shandong Province (No. ZR2015HQ018).
References
- 1.Mackintosh C, Madoz-Gúrpide J, Ordóñez JL, Osuna D, Herrero-Martín D. The molecular pathogenesis of Ewing's sarcoma. Cancer Biol Ther. 2010;9:655–667. doi: 10.4161/cbt.9.9.11511. [DOI] [PubMed] [Google Scholar]
- 2.Burningham Z, Hashibe M, Spector L, Schiffman JD. The epidemiology of sarcoma. Clin Sarcoma Res. 2012;2:14. doi: 10.1186/2045-3329-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, Kovar H, Joubert I, de Jong P, Rouleau G, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka K, Iwakuma T, Harimaya K, Sato H, Iwamoto Y. EWS-Fli1 antisense oligodeoxynucleotide inhibits proliferation of human Ewing's sarcoma and primitive neuroectodermal tumor cells. J Clin Invest. 1997;99:239–247. doi: 10.1172/JCI119152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joo J, Christensen L, Warner K, States L, Kang HG, Vo K, Lawlor ER, May WA. GLI1 is a central mediator of EWS/FLI1 signaling in Ewing tumors. PLoS One. 2009;4:e7608. doi: 10.1371/journal.pone.0007608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mateo-Lozano S, Gokhale PC, Soldatenkov VA, Dritschilo A, Tirado OM, Notario V. Combined transcriptional and translational targeting of EWS/FLI-1 in Ewing's sarcoma. Clin Cancer Res. 2006;12:6781–6790. doi: 10.1158/1078-0432.CCR-06-0609. [DOI] [PubMed] [Google Scholar]
- 7.Mortimore GE, Pösö AR, Lardeux BR. Mechanism and regulation of protein degradation in liver. Diabetes Metab Rev. 1989;5:49–70. doi: 10.1002/dmr.5610050105. [DOI] [PubMed] [Google Scholar]
- 8.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mortimore GE, Lardeux BR, Heydrick SJ. Mechanism and control of protein and RNA degradation in the rat hepatocyte: Two modes of autophagic sequestration. Revis Biol Celular. 1989;20:79–96. [PubMed] [Google Scholar]
- 10.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Qin ZH. Coordination of autophagy with other cellular activities. Acta Pharmacol Sin. 2013;34:585–594. doi: 10.1038/aps.2012.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deretic V. Autophagy in infection. Curr Opin Cell Biol. 2010;22:252–262. doi: 10.1016/j.ceb.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang S, Dupont N, Castillo EF, Deretic V. Secretory versus degradative autophagy: Unconventional secretion of inflammatory mediators. J Innate Immun. 2013;5:471–479. doi: 10.1159/000346707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janda E, Isidoro C, Carresi C, Mollace V. Defective autophagy in Parkinson's disease: Role of oxidative stress. Mol Neurobiol. 2012;46:639–661. doi: 10.1007/s12035-012-8318-1. [DOI] [PubMed] [Google Scholar]
- 15.Ren SY, Xu X. Role of autophagy in metabolic syndrome-associated heart disease. Biochim Biophys Acta. 2015;1852:225–231. doi: 10.1016/j.bbadis.2014.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi KS. Autophagy and cancer. Exp Mol Med. 2012;44:109–120. doi: 10.3858/emm.2012.44.2.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Czyzyk-Krzeska MF, Meller J, Plas DR. Not all autophagy is equal. Autophagy. 2012;8:1155–1156. doi: 10.4161/auto.20650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 19.Ogier-Denis E, Codogno P. Autophagy: A barrier or an adaptive response to cancer. Biochim Biophys Acta. 2003;1603:113–128. doi: 10.1016/s0304-419x(03)00004-0. [DOI] [PubMed] [Google Scholar]
- 20.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gélinas C, Fan Y, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ávalos Y, Canales J, Bravo-Sagua R, Criollo A, Lavandero S, Quest AF. Tumor suppression and promotion by autophagy. BioMed Res Int. 2014;2014:603980. doi: 10.1155/2014/603980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 23.Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. 'Protein modifications: Beyond the usual suspects' review series. EMBO Rep. 2008;9:859–864. doi: 10.1038/embor.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohsumi Y. Molecular dissection of autophagy: Two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 25.Tanida I, Ueno T, Kominami E. Human light chain 3/MAP1LC3B is cleaved at its carboxyl-terminal Met121 to expose Gly120 for lipidation and targeting to autophagosomal membranes. J Biol Chem. 2004;279:47704–47710. doi: 10.1074/jbc.M407016200. [DOI] [PubMed] [Google Scholar]
- 26.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 27.Tanida I, Sou YS, Ezaki J, Minematsu-Ikeguchi N, Ueno T, Kominami E. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J Biol Chem. 2004;279:36268–36276. doi: 10.1074/jbc.M401461200. [DOI] [PubMed] [Google Scholar]
- 28.Mariño G, Uría JA, Puente XS, Quesada V, Bordallo J, López-Otín C. Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. J Biol Chem. 2003;278:3671–3678. doi: 10.1074/jbc.M208247200. [DOI] [PubMed] [Google Scholar]
- 29.Hemelaar J, Lelyveld VS, Kessler BM, Ploegh HL. A single protease, Apg4B, is specific for the autophagy-related ubiquitin-like proteins GATE-16, MAP1-LC3, GABARAP, and Apg8L. J Biol Chem. 2003;278:51841–51850. doi: 10.1074/jbc.M308762200. [DOI] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Guo L, Huang JX, Liu Y, Li X, Zhou SR, Qian SW, Liu Y, Zhu H, Huang HY, Dang YJ, et al. Transactivation of Atg4b by C/EBPβ promotes autophagy to facilitate adipogenesis. Mol Cell Biol. 2013;33:3180–3190. doi: 10.1128/MCB.00193-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He C, Zhu H, Zhang W, Okon I, Wang Q, Li H, Le YZ, Xie Z. 7-Ketocholesterol induces autophagy in vascular smooth muscle cells through Nox4 and Atg4B. Am J Pathol. 2013;183:626–637. doi: 10.1016/j.ajpath.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herman-Antosiewicz A, Johnson DE, Singh SV. Sulforaphane causes autophagy to inhibit release of cytochrome c and apoptosis in human prostate cancer cells. Cancer Res. 2006;66:5828–5835. doi: 10.1158/0008-5472.CAN-06-0139. [DOI] [PubMed] [Google Scholar]
- 34.Harhaji-Trajkovic L, Vilimanovich U, Kravic-Stevovic T, Bumbasirevic V, Trajkovic V. AMPK-mediated autophagy inhibits apoptosis in cisplatin-treated tumour cells. J Cell Mol Med. 2009;13:3644–3654. doi: 10.1111/j.1582-4934.2009.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cabrera S, Maciel M, Herrera I, Nava T, Vergara F, Gaxiola M, López-Otín C, Selman M, Pardo A. Essential role for the ATG4B protease and autophagy in bleomycin-induced pulmonary fibrosis. Autophagy. 2015;11:670–684. doi: 10.1080/15548627.2015.1034409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song L, Liu H, Ma L, Zhang X, Jiang Z, Jiang C. Inhibition of autophagy by 3-MA enhances endoplasmic reticulum stress-induced apoptosis in human nasopharyngeal carcinoma cells. Oncol Lett. 2013;6:1031–1038. doi: 10.3892/ol.2013.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ewing J. Classics in oncology. Diffuse endothelioma of bone. James Ewing. Proceedings of the New York Pathological Society, 1921. CA Cancer J Clin. 1972;22:95–98. doi: 10.3322/canjclin.22.2.95. [DOI] [PubMed] [Google Scholar]
- 38.Stahl M, Ranft A, Paulussen M, Bölling T, Vieth V, Bielack S, Görtitz I, Braun-Munzinger G, Hardes J, Jürgens H, et al. Risk of recurrence and survival after relapse in patients with Ewing sarcoma. Pediatr Blood Cancer. 2011;57:549–553. doi: 10.1002/pbc.23040. [DOI] [PubMed] [Google Scholar]
- 39.Liu L, Yang M, Kang R, Wang Z, Zhao Y, Yu Y, Xie M, Yin X, Livesey KM, Lotze MT, et al. HMGB1-induced autophagy promotes chemotherapy resistance in leukemia cells. Leukemia. 2011;25:23–31. doi: 10.1038/leu.2010.225. [DOI] [PubMed] [Google Scholar]
- 40.Thompson AD, Teitell MA, Arvand A, Denny CT. Divergent Ewing's sarcoma EWS/ETS fusions confer a common tumorigenic phenotype on NIH3T3 cells. Oncogene. 1999;18:5506–5513. doi: 10.1038/sj.onc.1202928. [DOI] [PubMed] [Google Scholar]
- 41.Ma XH, Piao S, Wang D, McAfee QW, Nathanson KL, Lum JJ, Li LZ, Amaravadi RK. Measurements of tumor cell autophagy predict invasiveness, resistance to chemotherapy, and survival in melanoma. Clin Cancer Res. 2011;17:3478–3489. doi: 10.1158/1078-0432.CCR-10-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akin D, Wang SK, Habibzadegah-Tari P, Law B, Ostrov D, Li M, Yin XM, Kim JS, Horenstein N, Dunn WA., Jr A novel ATG4B antagonist inhibits autophagy and has a negative impact on osteosarcoma tumors. Autophagy. 2014;10:2021–2035. doi: 10.4161/auto.32229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu PF, Leung CM, Chang YH, Cheng JS, Chen JJ, Weng CJ, Tsai KW, Hsu CJ, Liu YC, Hsu PC, et al. ATG4B promotes colorectal cancer growth independent of autophagic flux. Autophagy. 2014;10:1454–1465. doi: 10.4161/auto.29556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rothe K, Lin H, Lin KB, Leung A, Wang HM, Malekesmaeili M, Brinkman RR, Forrest DL, Gorski SM, Jiang X. The core autophagy protein ATG4B is a potential biomarker and therapeutic target in CML stem/progenitor cells. Blood. 2014;123:3622–3634. doi: 10.1182/blood-2013-07-516807. [DOI] [PubMed] [Google Scholar]
- 45.Gump JM, Thorburn A. Autophagy and apoptosis: What is the connection? Trends Cell Biol. 2011;21:387–392. doi: 10.1016/j.tcb.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boya P, González-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, Métivier D, Meley D, Souquere S, Yoshimori T, et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thorburn J, Andrysik Z, Staskiewicz L, Gump J, Maycotte P, Oberst A, Green DR, Espinosa JM, Thorburn A. Autophagy controls the kinetics and extent of mitochondrial apoptosis by regulating PUMA levels. Cell Rep. 2014;7:45–52. doi: 10.1016/j.celrep.2014.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]