Figure 1.
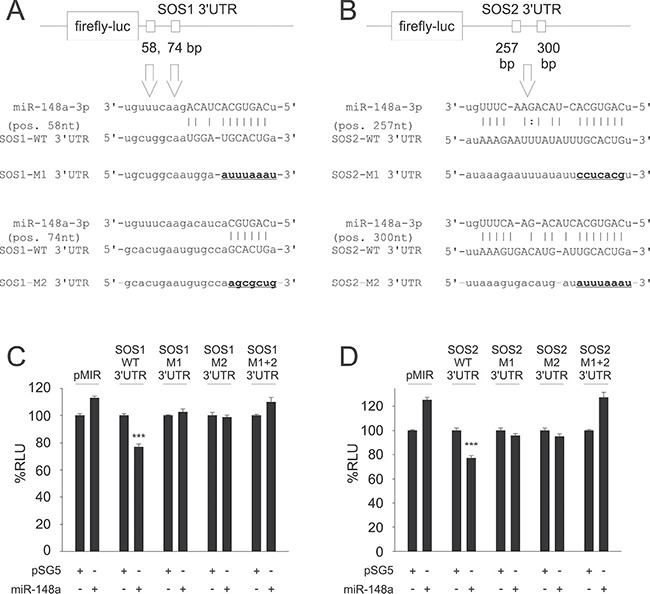
Predictions of potential binding sites for hsa-miR-148a-3p within the 3′UTRs of SOS1 (A) and SOS2 (B) and Dual-luciferase reporter assays for SOS1 (C) and SOS2 (D) as targets for miR-148a. Upper panels: Target site predictions. Annealing nucleotides between miR-148a-3p and its wildtype (WT) binding sites in SOS1 (A) and SOS2 (B) 3′UTRs are represented in upper case letters. Site directed mutagenesis of binding sites for miR-148a-3p (M1, M2) is indicated by underlined nucleotides in bold. Lower panels: Dual-luciferase assays. A pSG5 plasmid expressing miR-148a (or the empty pSG5 vector as a control) was co-transfected into HEK-293T cells either with the empty pMIR reporter vector, with pMIR reporter vector containing SOS1 (C) or SOS2 (D) wildtype (WT) 3′UTR, with SOS1 or SOS2 reporter vectors containing a single mutated binding site each (M1, M2) or with pMIR reporter vector for SOS1 (C) or SOS2 (D) containing both mutated binding sites (M1+2) each for miR-148a-3p. Co-transfection of miR-148a with wildtype 3′UTR reporter vectors resulted in relative light units significantly reduced to 77% for both SOS1 (C) or SOS2 (D) reporters as compared to control samples (P < 0.001 each). The downregulation of luciferase activity was abrogated as soon as at least one of the two miR-148a-3p binding sites was mutated.
