Figure 1. Method overview.
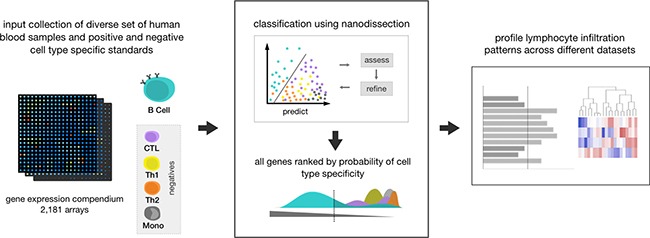
This diagram shows the process of running our method on one cell type, B cells. First all input data, consisting of over 2,000 human blood microarray samples taken in different conditions and different degrees of resolution, and positive and negative training standards, are assembled. In this example, B cell genes from Gene2MeSH are used as a positive standard and all other lymphocyte cell types (CTL, Th1, Th2, monocytes) in Gene2MeSH are used as negatives. Using the input, nanodissection finds a path that separates the positive training class from the negative ones in the space of the compendium, iteratively improving classification by selecting the best set of standards. Using the classification boundary, all genes assayed are then ranked by their probability to be cell type specific, in this case B cell specific. We then use the top 100 most probable cell type specific genes to estimate lymphocyte infiltration levels in breast cancer datasets.
