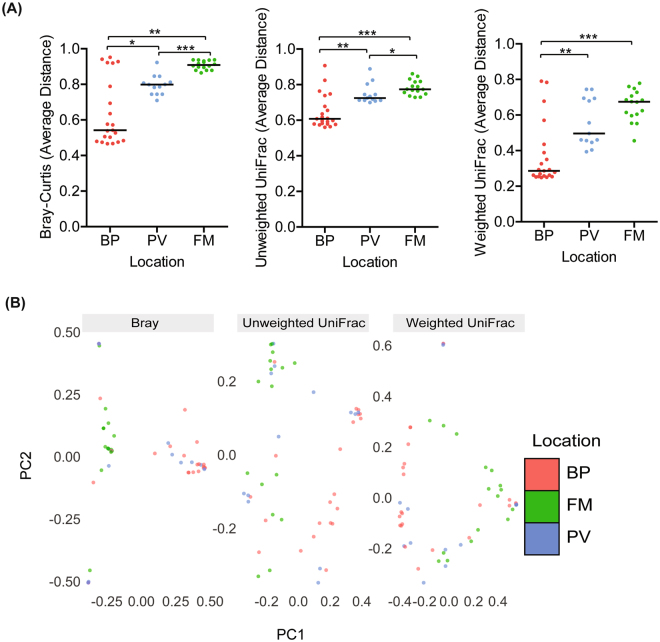
Figure 2.
Beta-diversity analysis illustrates greater similarity of microbial profiles within the basal plate samples than in samples from fetal membranes. (A) QIIME generated Bray-Curtis, unweighted Unifrac, and weighted UniFrac of average distances of validated samples grouped by sampling location. By all metrics, samples within the basal plate were significantly more similar to one another compared to samples within the placental villous or fetal membrane based on the median distance within each group (BP vs. PV–Bray-Curtis: P = 0.018; unweighted UniFrac: P = 0.0029; weighted UniFrac: P = 0.0011; BP vs. FM–Bray-Curtis: P = 0.0014; unweighted UniFrac: P = <0.0001; weighted UniFrac: P = 0.0002). Samples in the placental villous were significantly more similar to each other than samples within the fetal membrane (PV vs FM:–Bray-Curtis: P = <0.0001; unweighted UniFrac: P = 0.015; weighted UniFrac: P = 0.083). (B) Multidimensional scaling (MDS) plot of samples within the BP, PV, and FM based on the Bray-Curtis, unweighted UniFrac, and weighted UniFrac distance metrics (PERMANOVA: Bray: P = 0.001, weighted UniFrac: P = 0.001, weighted UniFrac: P = 0.001). Statistical analyses were performed using Mann-Whitney and PERMANOVA, and significant differences are designated with the following annotations: P = *<0.05, **<0.005, ***<0.0005.