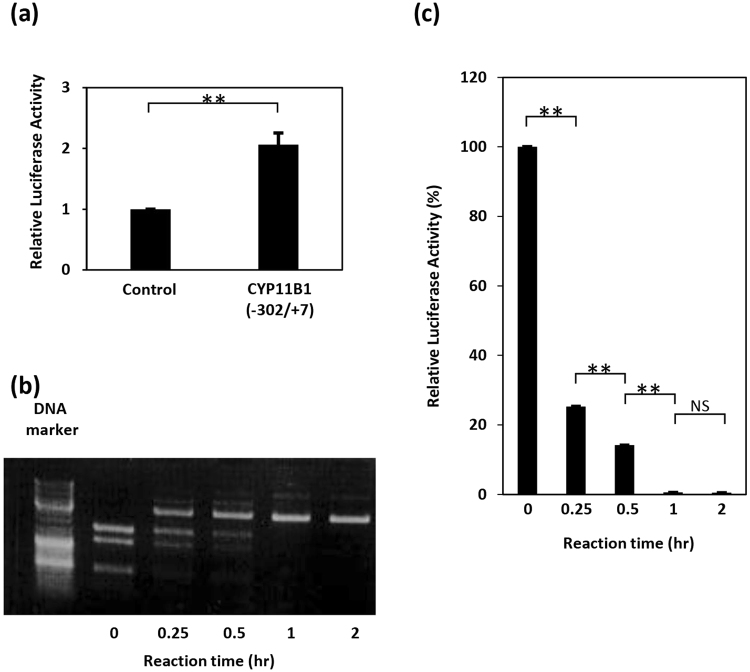
Figure 4.
DNA methylation suppresses CYP11B1 promoter activity. (a) Confirmation of CYP11B1 promoter activity. H295R cells were transiently transfected with pGL4.10[luc2] (control) or pGL4-cyp11b1[−302/+7](CYP11B1(−302/+7)). Two days after transfection, cells were lysed and luciferase activity was measured. The luciferase activity of the control sample was set to 1.0, and data are shown as the mean ± SEM (n = 6), and analyzed with the Mann-Whitney U test. **P < 0.01. (b) Confirmation of plasmid methylation. The CYP11B1 reporter plasmids were incubated with the CpG methyltransferase M.SssI for the indicated periods. The methylated plasmids were then digested by the methylation-sensitive restriction enzyme BsiEI and subjected to agarose electrophoresis. Note that BsiE1 cannot digest methylated plasmids. Data shown are representative of three independent experiments. Full-length gel is presented in Supplementary Figure S9. (c) Methylation suppresses CYP11B1 promoter activity. After incubation with M.SssI for the indicated periods, the CYP11B1 reporter plasmids were transfected into H295R cells, whose lysates were subjected to luciferase assay. Luciferase activity of the control sample (reaction time = 0 h) was set to 100%, and the mean ± SEM (n = 4) of the data are shown. Data are analyzed with the Mann-Whitney U test. *P < 0.05. NS, not significant.