Fig. 3.
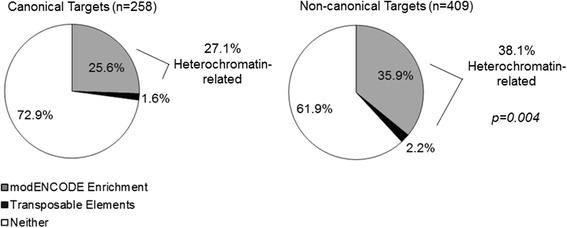
Non-canonical target sites have significantly more overlaps with heterochromatin sites. For both embryonic and maternal transcripts, overlaps between genomic loci corresponding to the probe set annotation and HP1a, Su(var)3–9 and/or H3K9me3 enriched loci listed in the relevant modENCODE database were tabulated. Probe sets annotated as transposable elements were also considered as heterochromatin sites. Non-canonical target probe sets had significantly higher proportion of such heterochromatin-related sites compared to canonical targets (p = 0.004, Fisher’s exact two-tailed test)
