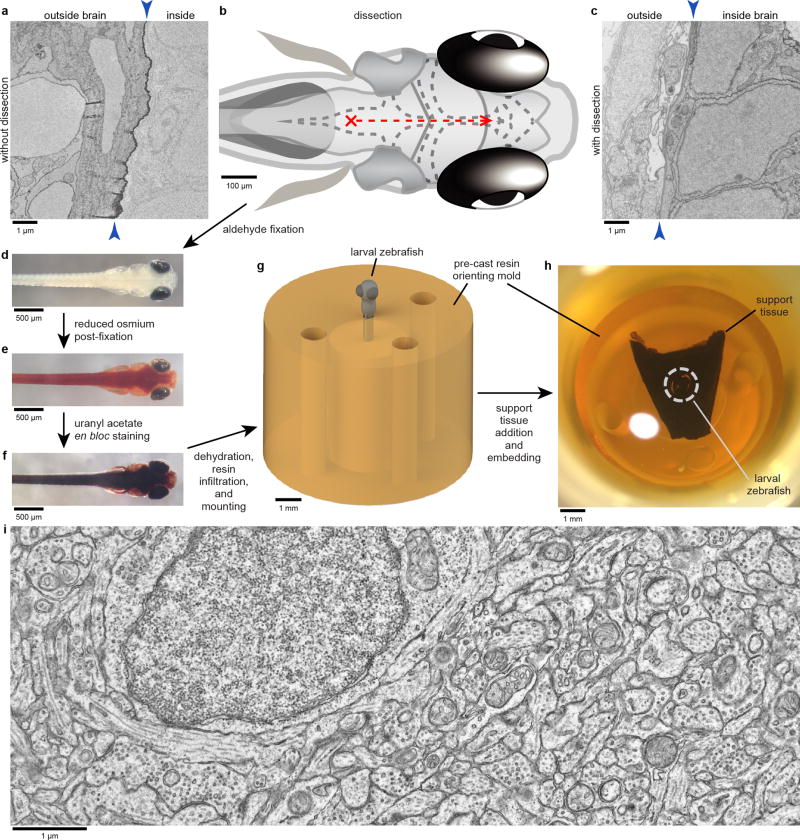
Extended Data Figure 1. Preparing larval zebrafish brain tissue for ssEM.
a, Immersion of intact specimens into tissue processing solutions resulted in poor preservation of brain ultrastructure due to membranes (arrowheads). b–c, Dissecting away the skin and membranes allowed solutions to diffuse into the brain, resulting in improved preservation. To minimize damage, dissections were initiated by puncturing the rhombencephalic ventricle dorsal to the hindbrain with a sharpened tungsten needle (red cross). Small anterior-directed incisions along the midline were then made as close to the surface as possible until the brain up to the anterior optic tectum was exposed (red dashed line). d–f, Following dissection and aldehyde fixation (d), samples were post-fixed with a reduced osmium solution (e) and stained with uranyl acetate (f). g–h, Processed specimens were then dehydrated with acetonitrile, infiltrated with a low-viscosity resin, mounted in a micromachined pre-cast resin mould to orient the sample for transverse sectioning (g), and surrounded by support tissue that stabilized sectioning (h). i, Representative ultrastructure acquired as a transmission electron micrograph from a section through the optic tectum of an early dissection test specimen. Scale bars: g–h, 1 mm; d–f, 500 µm; b, 100 µm; a,c,i, 1 µm.