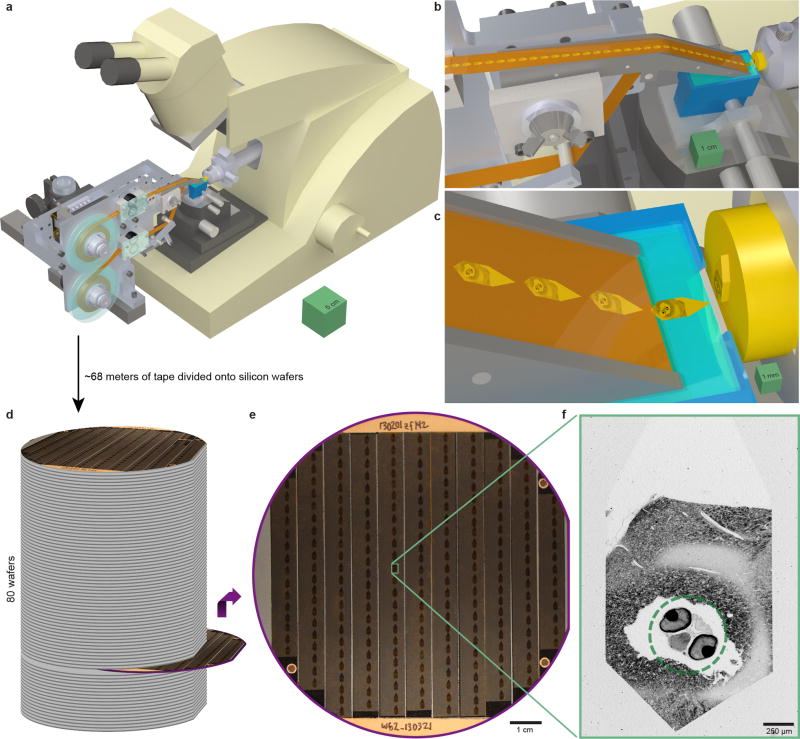
Extended Data Figure 2. Serial sectioning and ultrathin section library assembly.
a, Serial sections of resin-embedded samples were picked up with an automated tape-collecting ultramicrotome modified for compatibility with larger reels containing enough tape to accommodate tens of thousands of sections. b–c, Direct-to-tape sectioning resulted in consistent section spacing and orientation. Just as a section left the diamond knife (blue), it was caught by the tape. d, After sectioning, the tape was divided onto silicon wafers that functioned as a stage in a scanning electron microscope and formed an ultrathin section library. For a series containing all of a 5.5 dpf larval zebrafish brain, ~68 m of tape was divided onto 80 wafers (~227 sections per wafer). e, Wafer images were used as a coarse guide for targeting electron microscopic imaging. Fiducial markers (copper circles) further provided a reference for a per-wafer coordinate system, enabling storage of the position associated with each section for multiple rounds of re-imaging at varying resolutions as needed. f, 758.8×758.8×60 nm3vx−1 overview micrographs were acquired for each section to ascertain sectioning reliability and determine the extents of the ultrathin section library. Scale boxes: a, 5×5×5 cm3; b, 1×1×1 cm3; c, 1×1×1 mm3. Scale bars: e, 1 cm; f, 250 µm.