Fig. 5.
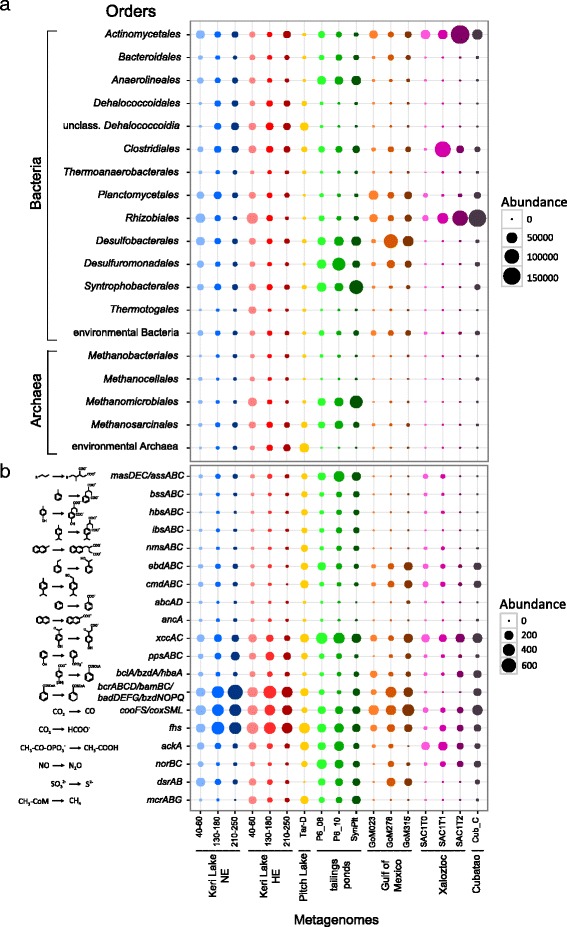
Comparison of Keri Lake metagenomes to publicly available datasets. a Abundances of selected prokaryotic orders in Keri Lake and publicly available metagenomes. b Absolute abundances of selected genes coding for enzymes involved in the pathways of anaerobic degradation of alkanes and aromatic compounds, Wood-Ljungdahl pathway, denitrification, sulfate reduction, and methanogenesis in Keri Lake and publicly available metagenomes
