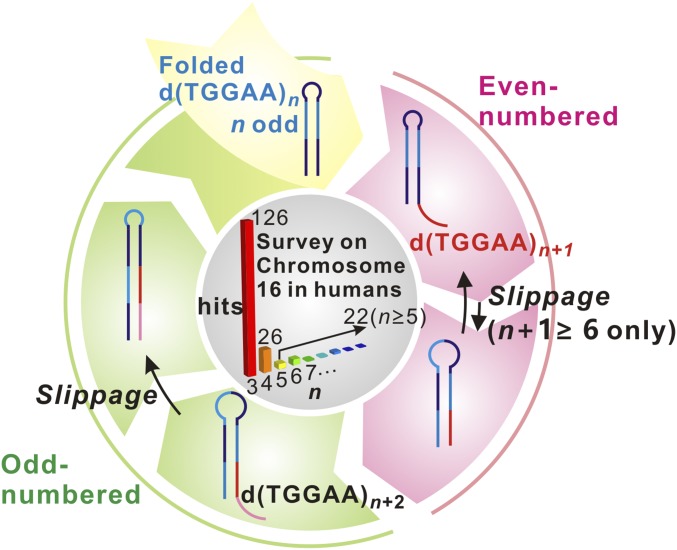
Fig. 6.
Proposed model for consecutive expansion of DNA repeats. From top and clockwise, initially odd-numbered d(TGGAA)n DNA folds into a hairpin structure, which has a high repeat expansion propensity. Expansion of one pentanucleotide unit dTGGAA results in a hairpin with an overhang product d(TGGAA)n+1. This product can undergo slippage motion and convert to an end-to-end aligned hairpin with an octanucleotide loop only if the repeat number n + 1 is greater than or equal to 6. Further expansion of the end-to-end aligned d(TGGAA)n+1 results in an odd-numbered d(TGGAA)n+2 product which has forms a hairpin structure again. The cycle can be perpetuated to yield long repeat expansions. A bioinformatics survey of the prevalence of each repeat number (n) in human chromosome 16 is shown in the center. The number of occurrences is shown at the top of the bars.