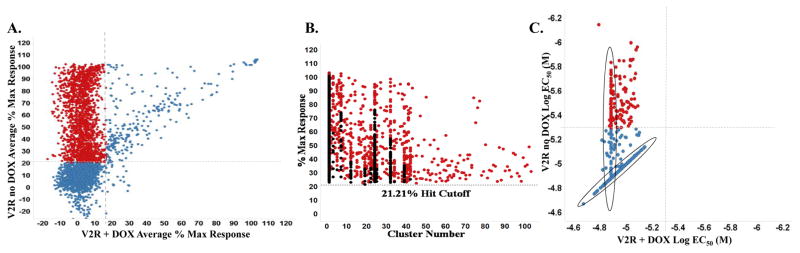
Figure 3.
A. Correlation plot of the V2R confirmation and counter screen data analysis of 3,734 compounds tested in triplicate at ~5.22μM plotted as the average % Max Response for the counterscreen on the X-axis (V2R+ DOX) vs. the confirmation assay on the Y-axis (V2R no DOX). Respective hit cutoffs are represented as dashed lines. Active and confirmed compounds that are not hits in the counterscreen are shown as red dots; unconfirmed or active also in the counterscreen are shown in blue. B. 1,694 compounds active only in V2R the “no DOX” assay format (red dots). These were clustered based on maximum common sub-structure similarity and ranked across all clusters to identify the top 640 most active and diverse compounds; highlighted as red dots. C. Correlation plot of primary and counterscreen EC50 data for 664 compounds tested in 10 point 1:3 dilution titration assays. Dashed lines represent activity cutoff parameters for each assay tested. Selectivity toward the primary V2R assay is demonstrated (red dots). Some compounds (circled) do not reach EC50 even at highest concentration tested (nominally 13.4μM).