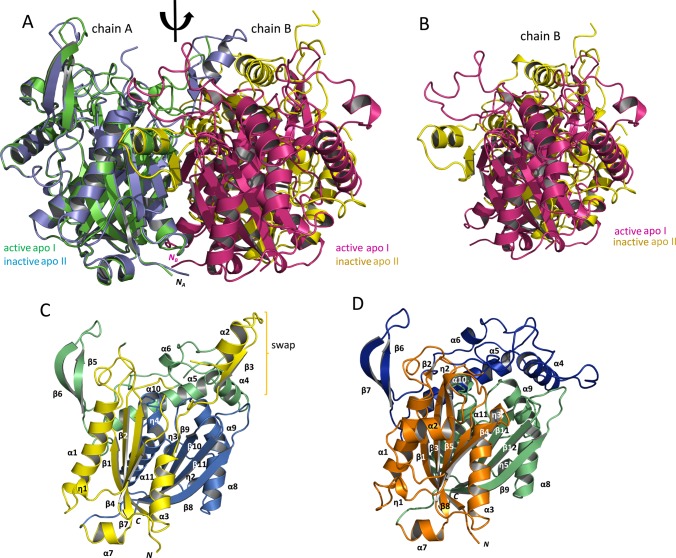
Figure 3.
Comparison of the FadA5 apo structures and illustration of the domain architecture of the oxidized enzyme. (A) Superposition of the two apo structure dimers, apo I (PDB entry 4UBW) with chain A, green, chain B, magenta; apo II (this publication) with chain A, blue, chain B, yellow in cartoon presentation. The monomers of apo I are related to each other by the indicated 2-fold axis. A conversion between the monomers of the apo II structure by the same axis is not possible. The N-termini of the monomers are indicated except for the N-terminus of the B chain within the apo II structure since it is not visible in this orientation. (B) Position of the two B chains resulting from the superposition of the A chains. The magenta (apo I) and the yellow (apo II) monomer are not congruently superimposed when A chains are aligned. They are in a different orientation with respect to monomer A, and thereby, the proteins form two different dimer interfaces. (C) The subdomain structure of the oxidized FadA5 apo II monomer is shown as a cartoon, consisting of the N-terminal subdomain (I, yellow), the lid subdomain (II, green), and the C-terminal subdomain (III, blue). The brackets highlight the swapped domain consisting of the α2-turn-ß3 motif (η = 310helix). (D) The subdomain structure of the apo I monomer is shown as a cartoon, consisting of the N-terminal subdomain (I, orange), the lid subdomain (II, blue), and the C-terminal subdomain (III, green). The monomer orientation is chosen according to panel C. Due to this orientation, η4, η6, and ß10 are not visible. The swapped domains here comprise the α2-turn-ß4 motif (η = 310helix).