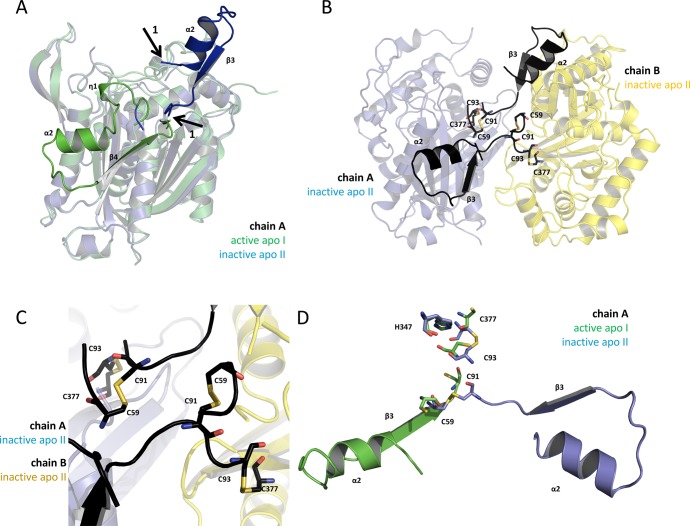
Figure 4.
The oxidized structure shows a swapped domain caused by the formation of a disulfide bond. (A) Superposition of the apo structures’ A chains (apo I, green; apo II; blue) is shown in transparent cartoon presentation. According to our activity data, the green monomer is active, and the blue monomer is inactive. The nontransparent secondary structure motifs, η1-α2-turn-ß4 (green) and α2-turn-ß3 (blue), are the regions of FadA5 which are swapped when comparing the apo I (active) with the apo II (inactive) protein structure. Arrows 1 indicate the starting and the end points of the reoriented domain (blue). The green domain is folding back to chain A, whereas the blue domain is pointing away from its monomer and forms an additional interface with chain B of the here described new apo II structure. (B) The apo II dimer is shown in cartoon presentation with the swapped domains being highlighted in black. The cysteines, which are forming the disulfide bonds (C59–C91 and C93–C377) in each monomer, are shown in stick presentation. Chain A is shown as transparent blue and chain B as transparent yellow cartoon. (C) Zoom of (B) to the residues that are forming the disulfide bonds in each monomer. (D) Superposition of the catalytic residues and the disulfide bond forming cysteines of the apo structures I (active, green) and II (inactive, blue) from chain A in stick presentation in the respective color code. The swapped domains of the apo II (inactive, blue) and apo I (active, green) structure of chain A are shown.