Abstract
Plants are constantly exposed to a variety of abiotic stresses, such as drought, heat, cold, flood, and salinity. To survive under such unfavorable conditions, plants have evolutionarily developed their own resistant-mechanisms. For several decades, many studies have clarified specific stress response pathways of plants through various molecular and genetic studies. In particular, it was recently discovered that ubiquitin proteasome system (UPS), a regulatory mechanism for protein turn over, is greatly involved in the stress responsive pathways. In the UPS, many E3 ligases play key roles in recognizing and tethering poly-ubiquitins on target proteins for subsequent degradation by the 26S proteasome. Here we discuss the roles of RING ligases that have been defined in related to abiotic stress responses in plants.
Keywords: Abiotic stress, RING E3 ligase, Ubiquitin proteasome system (UPS), Ubiquitin, 26s proteasome
INTRODUCTION
In eukaryotic cells, the ubiquitin proteasome system (UPS) determines protein turnover in response to various external stimuli, thereby regulates the cells’ ability to maintain their basal functions. In plants, UPS is an important part of defense against environmental stresses such as drought, extreme temperature, salinity, and infection by pathogenic micro-organisms (1–3). Many E3 ligases play a fundamental role in UPS, in which they conjugate target proteins with ubiquitins. The process mainly involves three steps. First, the ubiquitins are activated by an E1 activating enzyme, then transferred to an ubiquitin conjugating E2 enzyme and finally attached to the substrate protein by a specific ubiquitin E3 ligase. Then, the conjugated proteins become visible to the 26S proteasome which is responsible for the protein degradation process (4, 5). The importance of the protein turnover system is well exemplified in Arabidopsis, where about 6% of the Arabidopsis genome or about 1.600 genes encode core components of the UPS, including two E1s, at least 37 E2s and approximately 1.400 potential E3s (6). In particular, the E3 ligases are classified into four major different groups-the HECT (HOMOLOGY TO E6-AP C-TERMINUS)-type, RING (REALLY INTERESTING NEW GENE)-type, U-box-type and multi-complex E3 ligases - according to the type of their functional domains (4). HECT E3 ligases have a domain that is comprised of a conserved ~350 amino acids known as HECT domain. Using a cysteine residue located within the HECT domain, HECT E3 ligases form a thiol-ester intermediate with ubiquitin prior to tethering the ubiquitin to a target protein (7). In Arabidopsis, only seven different HECT E3 ligases have been defined and known as UBIQUITIN–PROTEIN LIGASES (UPLs), and they have been grouped into four subfamilies (UPL1/2, UPL3/4, UPL5, and UPL6/7) (7, 8). The RING-type E3s are characterized by the presence of a RING motif that coordinates two zinc ions to create a platform for binding of E2 enzymes. Over 470 genes of RING E3 ligases have been defined and classified into eight different classes according to the type of metal ligand residues (9) in Arabidopsis. Because of the large number of RING E3 ligases and their distinctive functions in diverse plant developmental processes and stress responses, an increasing number of studies on RING E3 ligases have been done in recent years. U-box E3 ligases are the most recently discovered class of E3 ligases that contain a conserved ~70 amino acids U-box motif (10, 11). U-box motif is a type of modified RING domain which lacks the zinc ion chelating residues (10, 11). In Arabidopsis, over 64 genes of U-box-type E3 ligases (Hereafter referred to as U-box) have been defined (12–14). These 64 U-box proteins were methodically named as PLANT U-box (PUB) with a serial number, except CARBOXYL TERMINUS OF HSC70-INTERACTING PROTEIN (CHIP). Recent studies on PUB genes have elucidated a wide range of functions of U-box E3 ligases in plant development and stress responses (12–14). In this mini review, with particular emphasis on the molecular and genetic analyses, we mainly aim to focus on the recent progress in understanding the functions of non-complex RING E3 ligases in abiotic stress responses.
NLA (NITROGEN LIMITATION ADAPTATION)
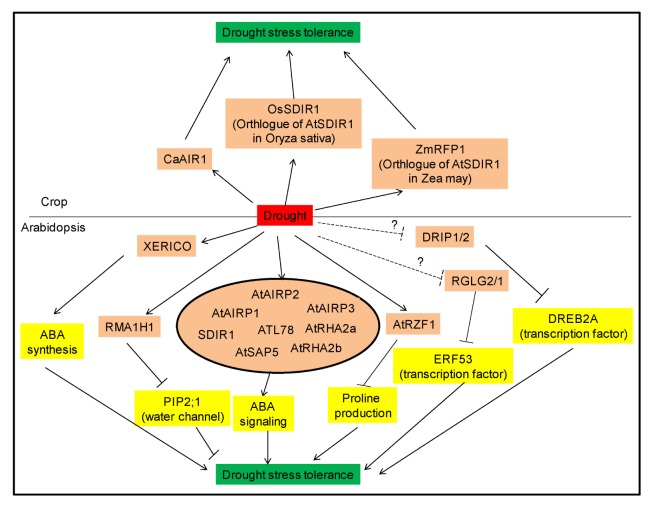
For the optimal growth and development of plants, nitrogen is required as an essential nutritional element, as can be seen by the growth defects that result from nitrogen deficiency. To overcome such abiotic stress, plants have evolved a set of adaptive responses to nitrogen-limiting stress. NLA (Nitrogen Limitation Adaptation), a gene that governs the adaptability of plants to nitrogen limitation, has been defined by a study. Under nitrogen-limiting condition, NLA-deficient plants fail to develop the essential responses that are needed for survival. Interestingly, NLA encodes a RING E3 ligase that localizes to the nucleus and directly interacts with Arabidopsis ubiquitin conjugase 8 (AtUBC8). In the absence of NLA activity, plants show premature senescence phenotype. Thus, NLA may be involved in the ubiquitination-mediated protein degradation of proteins as negative regulators in the nitrogen limitation sensing and signaling pathway (15).
Rma1H1 (RING MEMBRANE-ANCHOR 1)
Rma1H1, a hot pepper (Capsicum annuum) RING E3 ligase is homologous to a human RING membrane-anchor E3 ligase. Rma1H1 transcripts are immediately induced by various abiotic stresses, such as drought, high salinity, and cold. Also, overexpression of Rma1H1 in Arabidopsis conferred significantly enhanced tolerance to drought stress. Rma1H1 is able to tether poly-ubiquitins on itself and a target protein, which is an activity of an E3 ligase. Interestingly, Rma1H1 resides in the endoplasmic reticulum (ER) membrane and the overexpression of Rma1H1 hinders the trafficking of an aquaporin from the ER to the plasma membrane in Arabidopsis. Rma1H1 directly interacts with the aquaporin and guides the UPS-mediated degradation. Likewise, Rma1, an Arabidopsis homolog of Rma1H1, is also localized in the ER and is known to negatively regulate an aquaporin protein, PIP2:1. By blocking the trafficking of aquaporin to the plasma membrane from ER by proteolytic regulation, Rma1H1 and Rma1 play a critical role in response to dehydration in plants (16). Furthermore, Rma1H1-overexpressing tomato plants (35S:Rma1H1) show enhanced tolerance to high-salinity and drought when compared to wild-type plants. The overexpression of Rma1H1 leads to the dramatic up-regulation of ER chaperone genes such as LePDIL1, LeBIP1, and LeCNX1 in tomatoes. Therefore, overexpression of Rma1H1 may enhance the ER responses of tomato plants under drought stress not only by regulating aquaporin but also by effectively removing nonfunctional ubiquitinated proteins (17).
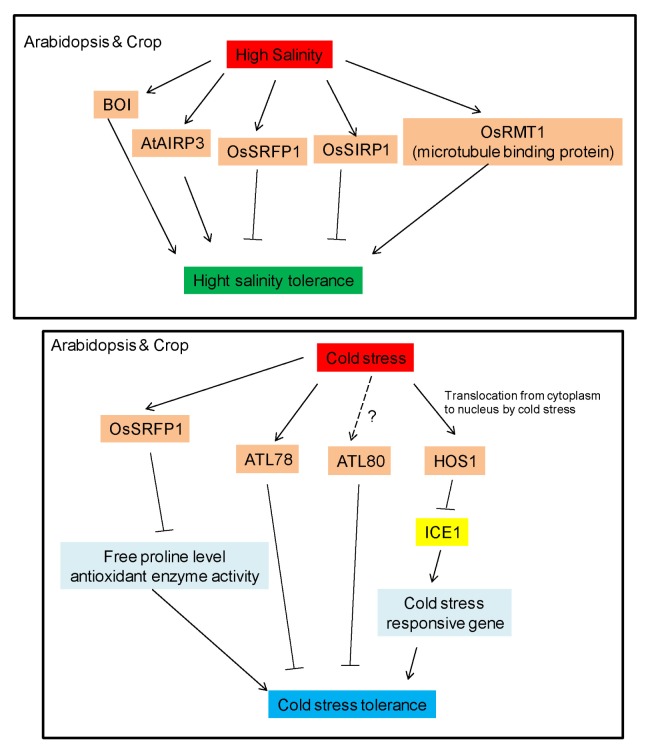
BOI (BOTRYTIS SUSCEPTIBLE INTERACTOR) and BRGs (BOI-RELATED GENES)
BOI (Botrytis Susceptible1 Interactor) directly interacts and ubiquitinates Arabidopsis BOS1, an R2R3MYB transcription factor which is involved in stress and pathogen responses. Indeed, BOI-knock down (RNAi) lines are known to be more susceptible to the necrotrophic fungus Botrytis cinerea and less tolerant to salt stress. Furthermore, three BOI-RELATED GENES (BRGs) were identified and have been known to contribute to B. cinerea resistance and salt stress responses. In fact, BOI RNAi and brg mutant plants show reduced growth in medium containing NaCl. The overexpression of BOI was less sensitive to salt compared to the wild-type plants. These results suggest that BOI and BRGs are involved in salt stress responses in Arabidopsis (18).
SDIR1 (SALT-AND DROUGHT-INDUCED RING FINGER 1)
In Arabidopsis, SALT-AND DROUGHT-INDUCED RING FINGER1 (SDIR1) is involved in abscisic acid (ABA)-related stress signal transduction (19). SDIR1 is notably up-regulated by drought and salt stress, but not by ABA. Overexpression of SDIR1 leads to ABA hypersensitivity and salt hypersensitivity in germination, enhanced stomatal closing and drought tolerance. The expression levels of a number of ABA-related and stress related genes are altered both in SDIR1-excesivity and sdir1-1 mutant plants. SDIR1 also seems to regulate leucine zipper family genes such as ABA-INSENSITIVE5 (ABI5), ABRE BINDING FACTOR3 (ABF3), and ABRE BINDING FACTOR4 (ABF4). In addition, the transcripts of OsSDIR1 (Oryza sativa SALT-AND DROUGHT-INDUCED RING FINGER 1), a homolog of SDIR1 in Arabidopsis, are up-regulated by drought and NaCl, but not by ABA. OsSDIR1 is able to complement the drought sensitive phenotype of the sdir1 mutant, showing that OsSDIR1 gene is functionally homologous to SDIR1. Upon drought treatment, the OsSDIR1-transgenic rice showed strong drought tolerance compared to control plants (20). ZmRFP1 is an ortholog of Arabidopsis SDIR1 gene in Zea may. The transcript levels of ZmRFP1 are markedly up-regulated by drought stress, and ABA treatment, but not by salt, heat and cold stresses (21). Although the gene expression profiles under different stresses vary in plants, these results suggest that SDIR1 plays diverse roles in ABA mediated salt and drought stress responses in plants (19–21).
AtRZF1 (ARABIDOPSIS THALIANA RING ZINC FINGER 1)
Arabidopsis RING Zinc Finger 1 (AtRZF1) is significantly reduced under drought stress. During early seedling development, atrzf1 mutant is less sensitive to osmotic stress than the wild-type. Besides, transgenic plants overexpressing AtRZF1 show various drought sensitive molecular and physiological phenotypes such as proline accumulation, water loss, membrane ion leakage and the expression of dehydration stress-related genes, showing that AtRZF1 negatively regulates early seedling development under drought stress (22). An ortholog of Arabidopsis thaliana RING Zinc Finger 1 (AtRZF1) gene was identified in the gourd family (Lagenaria siceraria) and was named LsRZF1. LsRZF1 transcripts are down-regulated by ABA, osmotic, and drought stresses. Transgenic plants ectopically expressing LsRZF1 in Arabidopsis are hypersensitive to ABA and osmotic stress during early seedling development, showing that LsRZF1 is also a negative regulator of drought stress response. AtRZF1 and LsRZF1 seem to be functional in a similar pathway that controls proline metabolism under drought condition (23). Interestingly, AtRZF1-deficiency is suppressed by a mutation in proline content alterative 22 (pca22). During early seedling growth, pca22 mutant plants suppress the insensitivity of atrzf1 to dehydration and ABA. These results suggest that pca22 is a dominant suppressor mutant of atrzf1 in the abiotic stress response (24).
AtAIRPs (ABA-INSENSITIVE RING PROTEINS)
Arabidopsis ABA-insensitive RING protein 1 (AtAIRP1) encodes a cytosolic protein containing a single RING domain. AtAIRP1 transcripts are significantly induced by ABA and drought stress. atairp1 mutant displays ABA-insensitive phenotypes at the germination stage. Besides, AtAIRP1-overexpressing transgenic plants show hypersensitive phenotypes to exogenous ABA such as radicle emergence, cotyledon development, root elongation, and stomatal closure. AtAIRP1-excessivity led to strong tolerance, as opposed to atairp1, which was highly susceptible to severe drought stress. The levels of drought stress-related genes and basic leucine zipper transcription factor genes are highly also up-regulated in AtAIRP1-excessive lines in response to ABA. AtAIRP1 is a positive regulator of ABA-dependent drought response (25). Arabidopsis ABA-insensitive RING protein 2 (AtAIRP2) encodes a cytosolic RING E3 Ub ligase whose expression is notably increased by ABA and dehydration stress. AtAIRP2-overexpressing transgenic plants showed hypersensitivity to ABA, whereas atairp2 loss-of-function mutant plants exhibited hyposensitive phenotypes, such as seed germination, root growth, and stomatal movement. AtAIRP2-excessivity displays a high tolerance to severe drought stress, and AtAIRP2-deficient plants are more susceptible to water stress than wild-type plants are. Interestingly, the positive effects of AtAIRP2 on ABA-induced stress genes are related to SNF1-related protein kinases, which are key components of the ABA signaling pathway. Furthermore, AtAIRP1 is functionally combinatory with AtAIRP2 in drought stress responses. With these results (26), suggested that AtAIRP2 is involved in the positive regulation of ABA-dependent drought stress responses. A recent study further revealed the detailed mechanism of AtAIRP2 in the crosstalk with ATP1/SDIRIP1, which was reported to be a negative factor in ABA signaling and a target protein of the RING E3 ligase (27). AtAIRP2 directly interacts with ATP1/SDIRIP1 and conjugates poly ubiquitins on ATP1/SDIRIP1. The half-life of ATP1/SDIRIP1 is regulated by AtAIRP2. Further detailed genetic analyses showed that ATP1/SDIRIP1 acts downstream of AtAIRP2. AtAIRP2 and SDIR1 reciprocally complemented the ABA- and salt-insensitive germination phenotypes of sdir1 and atairp2 mutants, respectively, indicating their combinatory roles in seed germination. Subcellular localization and BiFC analyses showed that AtAIRP2 and ATP1/SDIRIP1 are co-localized to the cytosolic spherical body, which is analogous to JUNQ body in yeast and animals. The regulation of ATP1/SDIRIP1 by AtAIRP2 is critical for ABA and high salinity responses during germination in Arabidopsis (27). ABA-insensitive RING protein 3 (AtAIRP3) contains three distinctive domains; a single RING domain, a putative myristoylation site, and a domain associated with RING2 (DAR2) domain. Transcript levels of AtAIRP3 are up-regulated by drought, high salinity, and ABA, indicating that AtAIRP3 plays a role in abiotic stress responses. In fact, the atairp3 knockout mutant and AtAIRP3-RNAi knockdown transgenic plants exhibit defective phenotypes in ABA-mediated seed germination and stomata closure. Compared to wild-type plants, the suppression of AtAIRP3 resulted in hypersensitive phenotypes under high salinity and water deficit. These results suggest that Arabidopsis ABA-insensitive RING protein 3 (AtAIRP3) is a positive regulator of the ABA-mediated drought and salt stress tolerance mechanisms. AtAIRP3 tethers ubiquitins to RESPONSIVE TO DEHYDRATION 21 (RD21), which is known as a drought-inducible Cys proteinase of the papain family (28). Thus AtAIRP3 plays a dual function in ABA-mediated drought stress responses as well as in amino acid export pathway in Arabidopsis (29).
KEG (KEEP ON GOING)
KEEP ON GOING (KEG) is a RING E3 ligase that harbors a RING motif, ankyrin repeats, a kinase domain and 12 HERC2-like repeats. KEG-deficient homo lines display growth arrest immediately after germination, extreme sensitivity to the inhibitory effects of ABA, and hypersensitivity to exogenous glucose, suggesting that KEG is a negative regulator of ABA signaling. KEG also accumulates high levels of ABSCISIC ACID-INSENSITIVE5 (ABI5) without additional ABA treatment, and has been shown to interact with ABI5 in vitro. In addition, ABI5-deficiency suppresses the growth-arrest phenotype of keg mutant seedlings. These results indicate that ABI5 is a target of KEG RING E3 ligase. In the absence of stress, KEG plays a key role in the regulation of ABA signaling pathway by degrading ABI5 (30), while ABA promotes the self-ubiquitination and degradation of KEG to maintain the level of ABI5 (31). Furthermore, a recent study suggested that KEG targets ABI5 for degradation in the cytoplasm, thereby inhibiting the nuclear accumulation of ABI5 in the absence of ABA (32).
ATLs (ARABIDOPSIS TÓXICOS EN LEVADURAS)
Among approximately 470 RING E3 ligases, 80 genes of RING in Arabidopsis and 121 genes in rice belong to ATL family. About 90% of the ATL genes are intronless, implying ATL E3 ligases may have evolved as a functional module. Several ATL genes are involved in abiotic stress responses in plants (33). For instance, AtATL78 is an Arabidopsis RING E3 ubiquitin ligase that is localized at the plasma membrane. The AtATL78 transcripts are up-regulated by cold stress, while down regulated by drought stress. In fact, the RNAi mediated suppression of AtATL78 shows enhanced tolerance to cold stress but decreased tolerance to drought. Therefore, AtATL78 seems to be a negative regulator of cold stress response and a positive regulator of drought stress response in Arabidopsis (34). Furthermore, AtATL78 mediates the ABA-dependent stomatal closure. The stomatal closure is fully impaired in atatl78 mutant plants even in the presence of exogenous ABA and reactive oxygen species (ROS). Treatment of high concentrations of Ca2+, a down-stream signaling molecule of ABA signaling pathway, to atatl78 mutant plants results in successful closures of the pores. In addition, the possible role of AtATL78 in promoting ROS-mediated ABA signaling pathway during drought stress has been elucidated (35). The T-DNA mutant of AtATL43 also shows an ABA-insensitive phenotype, suggesting a role of this gene in the ABA response (33). The transcripts of AtATL80, a plasma membrane (PM)-localized RING E3 ligase, are increased by long-term low Pi (0–0.02 mM KH2PO4) conditions in Arabidopsis seedlings. Overexpression of AtATL80 in Arabidopsis seedlings increased phosphorus (P) accumulation in the shoots and reduced P-utilization efficiency (PUE) even under high Pi (1 mM KH2PO4) conditions, while the atatl80 mutant line displayed opposite phenotypic traits. Phenotypes showing significant sensitivity to cold stress have been observed in AtATL80-overexpressing lines, whereas phenotypes with increased tolerance to cold stress have been observed in the atatl80 mutant line, suggesting that AtATL80 is a negative regulator of cold stress response (36). Further, the transcript of GmRFP1, an ATL-like RING E3 ligase gene of Soy bean (Glycin max) that contains an N-terminal transmembrane domain, is up-regulated by ABA and salt stress, but down-regulated by cold and drought stresses (37).
DRIP1 (DREB2A-INTERACTING PROTEIN1)
DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN2A (DREB2A) is a transcription factor that controls water deficit-inducible gene expression. It was recently found that the stability of DREB2A in the nucleus is important for its activation, a process which requires posttranslational modification. Arabidopsis thaliana DREB2A-INTERACTING PROTEIN1 (DRIP1) and DRIP2, are RING E3 ligases that directly interact and tether ubiquitins to DREB2A protein in the nucleus. The expressions of DREB2A-regulated drought-responsive genes are negatively regulated by DRIP1. By contrast, the expressions of the drought-inducible genes are slightly increased in the single T-DNA mutants of drip1-1 and drip2-1. Notably, under dehydration stress, the gene expressions are significantly enhanced in the drip1/drip2 double mutant, implying that DRIP1 and DRIP2 function negatively in response to drought stress by targeting DREB2A in plants (38). An ortholog of DRIP was found in Cowpea (Vigna unguiculata L. Walp) (VuDRIP) using PCR based methods. VuDRIP transcripts are up-regulated in response to various abiotic stresses and phytohormones. In addition, VuDRIP also binds to VuDREB2A in yeast two-hybrid assay. The results indicate the negative regulation of VuDREB2A by ubiquitin ligases in cowpea similar to Arabidopsis (39).
XERICO
XERICO encodes a relatively small RING E3 ligase with an N-terminal trans-membrane domain (162 aa), and its transcripts seem to be induced by salt and osmotic stress. During germination and early seedling growth, transgenic Arabidopsis plants overexpressing XERICO show hypersensitivity to salt and osmotic stress, as well as ABA treatment. Under drought stress, XERICO enhances the expression of AtNCED3, a key ABA-biosynthetic gene. Consistently, the upregulation of XERICO under the control of 35S promoter significantly accumulates cellular ABA levels, and also shows strong tolerance against drought stress. In addition, XERICO interacts with ASK1-interacting F-box protein (AtTLP9), which is involved in the ABA-signaling pathway. Furthermore, the expressions of many genes for plant hormones are notably altered by XERICO overexpression (40).
HOS1 (HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE)
High expression of osmotically responsive gene 1 (HOS1) is a RING E3 ligase that is generally expressed in all plant tissues. HOS1 negatively regulates low temperature-responsive gene transcription, and its deficiency results in the enhanced induction of the CBF transcription factors and their downstream cold-responsive genes by cold treatment (41). Interestingly, in response to low temperature treatments, HOS1 protein shuttles from the cytoplasm to the nucleus (42). Further studies elucidated the detailed mechanism of underlying the functions of HOS1 in cold stress response. Inducer of CBF expression 1 (ICE1), which is a transcription factor, activates the expression of C-repeat (CRT)-binding factors (CBFs) that subsequently controls the expression of downstream genes for cold responses (43). The direct interaction of HOS1 with ICE1 mediates the ubiquitination of ICE1, which in turn mediates the cold induced degradation of ICE1. Consistently, HOS1-deficiency leads to the accumulation of ICE1 and downstream genes, while the overexpression of HOS1 reduces the level of ICE1 by UPS mediated degradation. As such, HOS1 plays a key role in attenuating cold stress responses in Arabidopsis (44).
CaAIR1 (CAPSICUM ANNUUM ABA-INSENSITIVE RING PROTEIN 1)
Capsicum annuum ABA-Insensitive RING protein 1 (CaAIR1) is a RING E3 ligase for a hypersensitive response to drought stress in hot pepper (45). CaAIR1, which contains a RING domain and a putative transmembrane domain, has a RING E3 ligase activity in vitro and is localized in the nucleus. The transcripts of CaAIR1 are up-regulated by ABA treatments, drought and NaCl stresses, indicating its putative roles in the response to abiotic stress. The ectopic expression of CaAIR1 in Arabidopsis leads to an impaired sensitivity to ABA during plant developmental stages and hypersensitive drought stress phenotypes such as reduced stomatal closure, while CaAIR1-deficiency in pepper plants results in an enhanced tolerance to drought stress. These results indicate that CaAIR1 participates in the ABA-mediated drought stress tolerance mechanism as a negative regulator.
OsHCI1 (ORYZA SATIVA HEAT AND COLD INDUCED 1)
Oryza sativa heat and cold induced 1 (OsHCI1) is a RING E3 ligase gene which is highly up-regulated by heat and cold stress but not by salinity or dehydration. OsHCI1 associates with six target proteins such as 20S proteasome subunit α7 (OsPSA7, Os01g59600), periplasmic beta-glucosidase (OsBGLU1, Os03g53800), ethylene-responsive protein (OsbHLH065, Os04g41570), glycine-rich cell-wall structural protein (OsGRP1, Os05g02770), peroxidase (OsPOX1, Os07g48020), and 14-3-3 protein (Os14-3-3, Os11g34450). In particular, OsHCI1 mediates OsPGLU1, OsbHLH065, OsGRP1, and OsPOX1 protein ubiquitination in vitro. The mono-ubiquitin of the target proteins seem to be important for the subcellular trafficking of the proteins from the nucleus to the cytoplasm. Under heat shock conditions, OsHCI1 also dynamically moves from the cytoplasm to the nucleus along the cytoskeletal tracts. Furthermore, the ectopic expression of YFP fused OsHCI1 in Arabidopsis shows a heat-tolerant phenotype and increased survival rate under heat stress, indicating that OsHCI1 indeed plays an important role in the regulation of heat-generated signals in plants (46).
OsRMT1 (ORYZA SATIVA RING FINGER PROTEIN WITH MICROTUBULE-TARGETING DOMAIN 1)
Oryza sativa RING finger protein with microtubule-targeting domain 1 (OsRMT1), is a functional RING E3 ligase that is possibly involved in a salt tolerance mechanism. As a homodimer complex, OsRMT1 functions as a self-ubiquitination-mediated protein degradation under nonstress conditions. By contrast, under salt stress, OsRMT1 is stabilized and localized in the nucleus and microtubules. In fact, the N-terminal domain of OsRMT1 is required for microtubule targeting. The overexpression of OsRMT1 in Arabidopsis exhibits increased tolerance to salt stress, while the abundance of microtubule-associated OsRMT1 is strictly regulated under nonstress condition. Together, this indicates that OsRMT1 may play a role in salt stress response by modulating the half-life of target proteins that are mostly associated to microtubules (47).
OsSIRP1 (ORYZA SATIVA SALT-INDUCED RING FINGER PROTEIN 1)
Oryza sativa salt-induced RING Finger Protein 1 (OsSIRP1) gene was selected among the 44 Oryza sativa RING Finger Proteins (OsRFPs) genes which are highly expressed in rice roots under salinity stress. OsSIRP1 transcripts are up-regulated by various stress treatments, including salt, heat, drought and ABA. In vitro poly-ubiquitination assay showed that OsSIRP1 is indeed, a functional E3 ligase. The ectopic overexpression of OsSIRP1 in Arabidopsis reduces tolerance for salinity stress during seed germination and root growth. Thus, it has been elucidated OsSIRP1 may act as a negative regulator of salinity stress tolerance (48).
OsSRFP1 (ORYZA SATIVA STRESS-RELATED RING FINGER PROTEIN 1)
In vitro ubiquitination assay revealed that OsSRFP1 has E3 ubiquitin ligase activity. OsSRFP1 is ubiquitously expressed in various rice organs and is induced by cold, dehydration, salt, H2O2 and ABA treatments. The overexpression of OsSRFP1 reduces stress tolerance to salt, cold and oxidative stresses, whereas RNAi-knock down of OsSRFP1 increases tolerance against to those stresses. Consistently, the amount of free proline and the activities of antioxidant enzymes are increased in the RNAi-knock down lines but decreased in the over-expression lines under cold stress. Furthermore, OsSRFP1 may be functional in the regulation of several gene transcriptions. The expressions of many genes for reactive oxygen species (ROS) homeostasis are altered by OsSRFP1-excessivity under normal and cold conditions. By negatively regulating antioxidant enzymes for the removal of ROS, OsSRFP1 may have a dual function in post-translational and transcriptional regulations as well as in modulating abiotic stress responses in rice (49).
CONCLUSION AND PERSPECTIVES
As sessile organisms, plants are constantly subjected to numerous abiotic stresses residing in their immediate surroundings. Generally, plants overcome such adverse conditions altering their physiological, biochemical, and morphological phenotypes. A variety of plant adaptation pathways against harmful abiotic stresses have been investigated and depicted. Among such variety of pathways, UPS is one of the key processes through which plants are able to maintain growth against environmental stresses. Hence, it is of particular importance to further understand the roles of E3 ligases of the UPS in the process of plant adaptation against stress conditions. Through genomic and transcriptome analyses, a growing number of RING E3 ligases related to abiotic stress responses have been identified (Fig. 1). The roles of RING E3 ligases as either positive or negative regulators in response to stress mainly depend on the nature of their target proteins (Fig. 2). However, our efforts to understand the detailed regulatory roles of RING E3 ligases at the molecular and biochemical levels is often hindered by the lack of information on such target proteins. Therefore, the identification of target proteins is necessary to gain a better understanding of the biological functions of RING E3 ligases in abiotic stress responses. Although the target proteins of several abiotic stress related RING E3 ligases, such as HOS1, RmaH1, and DREB2A, have been defined using conventional molecular approaches, information on the target proteins of most RING E3 ligases remain elusive due to the lack of efficient screening system for substrates. The quest for future researches is to build a high-throughput method that can screen unknown targets proteins. With this method, the roles of RING E3 ligases and other families in not only plant abiotic stress management but also growth and development can be further understood. After establishing the functions of individual RING E3 ligases, the regulatory networks among the stress response pathways for different stresses should be interlinked to grasp in-depth knowledge on stress responses in plants. Moreover, contrary to yeast and animals, our knowledge on E3 ligases under proteotoxic stress is still relatively rudimentary in plants. In fact, only one U-box E3 ligase AtCHIP has been identified for protein quality control (PQC) pathway (50, 51). PQC is one of most important biological processes by which misfolded, damaged, and truncated proteins can be rescued or degraded to prevent unfavorable aberrant protein aggregations. Indeed, many intracellular proteins are prone to be damaged by many abiotic stresses, such as drought, heat, and oxidative stresses (52–54). Therefore, in relation to the PQC process in plants, the roles of E3 ligases in the abiotic stresses need further understanding. Given the high diversity of RING E3 ligases in plants, it seems reasonable to postulate the involvement of some of these RING E3 ligases in proteostasis. Thus in future perspective, it is of high interest to study and understand the possible genes that may be involved in proteostasis.
Fig. 1.
Drought stress related RING E3 ligases in plants. Schematic cartoon shows the positive or negative functions of RING E3 ligases.
Fig. 2.
Cold stress and High salinity related RING E3 ligases in plants. Schematic cartoon shows the positive or negative functions of RING E3 ligases.
ACKNOWLEDGEMENTS
This work was supported by grants from the National Center for Next-Generation BioGreen21 Program (PJ01194601) to S.W.Y., and the National Research Foundation (Project number: NRF-2016K2A9A1A06922119) to W.T.K.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting financial interests.
REFERENCES
- 1.Lyzenga WJ, Stone SL. Abiotic stress tolerance mediated by protein ubiquitination. J Exp Bot. 2012;63:599–616. doi: 10.1093/jxb/err310. [DOI] [PubMed] [Google Scholar]
- 2.Dreher K, Callis J. Ubiquitin, hormones and biotic stress in plants. Ann Bot. 2007;99:787–822. doi: 10.1093/aob/mcl255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belknap WR, Garbarino JE. The role of ubiquitin in plant senescence and stress responses. Trends Plant Sci. 1996;1:331–335. doi: 10.1016/S1360-1385(96)82593-0. [DOI] [Google Scholar]
- 4.Vierstra RD. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol. 2009;10:385–397. doi: 10.1038/nrm2688. [DOI] [PubMed] [Google Scholar]
- 5.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 6.Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol. 2004;55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- 7.Huibregtse JM, Scheffner M, Beaudenon S, Howley PM. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci U S A. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Downes BP, Stupar RM, Gingerich DJ, Vierstra RD. The HECT ubiquitin-protein ligase (UPL) family in Arabidopsis: UPL3 has a specific role in trichome development. Plant J. 2003;35:729–742. doi: 10.1046/j.1365-313X.2003.01844.x. [DOI] [PubMed] [Google Scholar]
- 9.Stone SL, Hauksdóttir H, Troy A, Herschleb J, Kraft E, Callis J. Functional Analysis of the RING-Type Ubiquitin Ligase Family of Arabidopsis. Plant Physiol. 2005;137:13–30. doi: 10.1104/pp.104.052423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohi MD, Vander Kooi CW, Rosenberg JA, Chazin WJ, Gould KL. Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat Struct Biol. 2003;10:250–255. doi: 10.1038/nsb906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aravind L, Koonin EV. The U box is a modified RING finger - a common domain in ubiquitination. Curr Biol. 2000;10:R132–134. doi: 10.1016/S0960-9822(00)00398-5. [DOI] [PubMed] [Google Scholar]
- 12.Yee D, Goring DR. The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. J Exp Bot. 2009;60:1109–1121. doi: 10.1093/jxb/ern369. [DOI] [PubMed] [Google Scholar]
- 13.Andersen P, Kragelund BB, Olsen AN, et al. Structure and biochemical function of a prototypical Arabidopsis U-box domain. J Biol Chem. 2004;279:40053–40061. doi: 10.1074/jbc.M405057200. [DOI] [PubMed] [Google Scholar]
- 14.Wiborg J, O’Shea C, Skriver K. Biochemical function of typical and variant Arabidopsis thaliana U-box E3 ubiquitin-protein ligases. Biochem J. 2008;413:447–457. doi: 10.1042/BJ20071568. [DOI] [PubMed] [Google Scholar]
- 15.Peng M, Hannam C, Gu H, Bi YM, Rothstein SJ. A mutation in NLA, which encodes a RING-type ubiquitin ligase, disrupts the adaptability of Arabidopsis to nitrogen limitation. Plant J. 2007;50:320–337. doi: 10.1111/j.1365-313X.2007.03050.x. [DOI] [PubMed] [Google Scholar]
- 16.Lee HK, Cho SK, Son O, Xu Z, Hwang I, Kim WT. Drought stress-induced Rma1H1, a RING membrane-anchor E3 ubiquitin ligase homolog, regulates aquaporin levels via ubiquitination in transgenic Arabidopsis plants. Plant Cell. 2009;21:622–641. doi: 10.1105/tpc.108.061994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seo YS, Choi JY, Kim SJ, Kim EY, Shin JS, Kim WT. Constitutive expression of CaRma1H1, a hot pepper ER-localized RING E3 ubiquitin ligase, increases tolerance to drought and salt stresses in transgenic tomato plants. Plant Cell Rep. 2012;31:1659–1665. doi: 10.1007/s00299-012-1278-0. [DOI] [PubMed] [Google Scholar]
- 18.Luo H, Laluk K, Lai Z, Veronese P, Song F, Mengiste T. The Arabidopsis Botrytis Susceptible1 Interactor defines a subclass of RING E3 ligases that regulate pathogen and stress responses. Plant Physiol. 2010;154:1766–1782. doi: 10.1104/pp.110.163915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Yang C, Li Y, et al. SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell. 2007;19:1912–1929. doi: 10.1105/tpc.106.048488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao T, Wu Y, Zhang Y, et al. OsSDIR1 overexpression greatly improves drought tolerance in transgenic rice. Plant Mol Biol. 2011;76:145–156. doi: 10.1007/s11103-011-9775-z. [DOI] [PubMed] [Google Scholar]
- 21.Xia Z, Liu Q, Wu J, Ding J. ZmRFP1, the putative ortholog of SDIR1, encodes a RING-H2 E3 ubiquitin ligase and responds to drought stress in an ABA-dependent manner in maize. Gene. 2012;495:146–153. doi: 10.1016/j.gene.2011.12.028. [DOI] [PubMed] [Google Scholar]
- 22.Ju HW, Min JH, Chung MS, Kim CS. The atrzf1 mutation of the novel RING-type E3 ubiquitin ligase increases proline contents and enhances drought tolerance in Arabidopsis. Plant Sci. 2013;203–204:1–7. doi: 10.1016/j.plantsci.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 23.Min J-H, Ju H-W, Yang K-Y, Chung J-S, Cho B-H, Kim CS. Heterologous expression of the gourd E3 ubiquitin ligase gene LsRZF1 compromises the drought stress tolerance in Arabidopsis thaliana. Plant Physiology and Biochemistry. 2014;77:7–14. doi: 10.1016/j.plaphy.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 24.Kim AR, Min JH, Lee KH, Kim CS. PCA22 acts as a suppressor of atrzf1 to mediate proline accumulation in response to abiotic stress in Arabidopsis. J Exp Bot. 2017;68:1797–1809. doi: 10.1093/jxb/erx069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryu MY, Cho SK, Kim WT. The Arabidopsis C3H2C3-type RING E3 ubiquitin ligase AtAIRP1 is a positive regulator of an abscisic acid-dependent response to drought stress. Plant Physiol. 2010;154:1983–1997. doi: 10.1104/pp.110.164749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho SK, Ryu MY, Seo DH, Kang BG, Kim WT. The Arabidopsis RING E3 ubiquitin ligase AtAIRP2 plays combinatory roles with AtAIRP1 in abscisic acid-mediated drought stress responses. Plant Physiol. 2011;157:2240–2257. doi: 10.1104/pp.111.185595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh TR, Kim JH, Cho SK, Ryu MY, Yang SW, Kim WT. AtAIRP2 E3 Ligase affects ABA and High Salinity Responses by Stimulating its ATP1/SDIRIP1 Substrate Turnover. Plant Physiol. 2017;174:2515–2531. doi: 10.1104/pp.17.00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koizumi M, Yamaguchi-Shinozaki K, Tsuji H, Shinozaki K. Structure and expression of two genes that encode distinct drought-inducible cysteine proteinases in Arabidopsis thaliana. Gene. 1993;129:175–182. doi: 10.1016/0378-1119(93)90266-6. [DOI] [PubMed] [Google Scholar]
- 29.Kim JH, Kim WT. The Arabidopsis RING E3 ubiquitin ligase AtAIRP3/LOG2 participates in positive regulation of high-salt and drought stress responses. Plant Physiol. 2013;162:1733–1749. doi: 10.1104/pp.113.220103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J. KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell. 2006;18:3415–3428. doi: 10.1105/tpc.106.046532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu H, Stone SL. Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell. 2010;22:2630–2641. doi: 10.1105/tpc.110.076075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu H, Stone SL. Cytoplasmic degradation of the Arabidopsis transcription factor abscisic acid insensitive 5 is mediated by the RING-type E3 ligase KEEP ON GOING. J Biol Chem. 2013;288:20267–20279. doi: 10.1074/jbc.M113.465369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Serrano M, Parra S, Alcaraz LD, Guzman P. The ATL gene family from Arabidopsis thaliana and Oryza sativa comprises a large number of putative ubiquitin ligases of the RING-H2 type. J Mol Evol. 2006;62:434–445. doi: 10.1007/s00239-005-0038-y. [DOI] [PubMed] [Google Scholar]
- 34.Kim SJ, Kim WT. Suppression of Arabidopsis RING E3 ubiquitin ligase AtATL78 increases tolerance to cold stress and decreases tolerance to drought stress. FEBS Lett. 2013;587:2584–2590. doi: 10.1016/j.febslet.2013.06.038. [DOI] [PubMed] [Google Scholar]
- 35.Suh JY, Kim SJ, Oh TR, Cho SK, Yang SW, Kim WT. Arabidopsis Toxicos en Levadura 78 (AtATL78) mediates ABA-dependent ROS signaling in response to drought stress. Biochem Biophys Res Commun. 2016;469:8–14. doi: 10.1016/j.bbrc.2015.11.061. [DOI] [PubMed] [Google Scholar]
- 36.Suh JY, Kim WT. Arabidopsis RING E3 ubiquitin ligase AtATL80 is negatively involved in phosphate mobilization and cold stress response in sufficient phosphate growth conditions. Biochem Biophys Res Commun. 2015;463:793–799. doi: 10.1016/j.bbrc.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 37.Du QL, Cui WZ, Zhang CH, Yu DY. GmRFP1 encodes a previously unknown RING-type E3 ubiquitin ligase in Soybean (Glycine max) Mol Biol Rep. 2010;37:685–693. doi: 10.1007/s11033-009-9535-1. [DOI] [PubMed] [Google Scholar]
- 38.Qin F, Sakuma Y, Tran LS, et al. Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell. 2008;20:1693–1707. doi: 10.1105/tpc.107.057380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sadhukhan A, Panda SK, Sahoo L. The cowpea RING ubiquitin ligase VuDRIP interacts with transcription factor VuDREB2A for regulating abiotic stress responses. Plant Physiol Biochem. 2014;83:51–56. doi: 10.1016/j.plaphy.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 40.Ko JH, Yang SH, Han KH. Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J. 2006;47:343–355. doi: 10.1111/j.1365-313X.2006.02782.x. [DOI] [PubMed] [Google Scholar]
- 41.Ishitani M, Xiong L, Lee H, Stevenson B, Zhu JK. HOS1, a genetic locus involved in cold-responsive gene expression in arabidopsis. Plant Cell. 1998;10:1151–1161. doi: 10.1105/tpc.10.7.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee H, Xiong L, Gong Z, Ishitani M, Stevenson B, Zhu JK. The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev. 2001;15:912–924. doi: 10.1101/gad.866801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee BH, Henderson DA, Zhu JK. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell. 2005;17:3155–3175. doi: 10.1105/tpc.105.035568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong CH, Agarwal M, Zhang Y, Xie Q, Zhu JK. The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc Natl Acad Sci U S A. 2006;103:8281–8286. doi: 10.1073/pnas.0602874103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park C, Lim CW, Baek W, Lee SC. RING Type E3 Ligase CaAIR1 in Pepper Acts in the Regulation of ABA Signaling and Drought Stress Response. Plant Cell Physiol. 2015;56:1808–1819. doi: 10.1093/pcp/pcv103. [DOI] [PubMed] [Google Scholar]
- 46.Lim SD, Cho HY, Park YC, Ham DJ, Lee JK, Jang CS. The rice RING finger E3 ligase, OsHCI1, drives nuclear export of multiple substrate proteins and its heterogeneous overexpression enhances acquired thermotolerance. J Exp Bot. 2013;64:2899–2914. doi: 10.1093/jxb/ert143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lim SD, Jung CG, Park YC, et al. Molecular dissection of a rice microtubule-associated RING finger protein and its potential role in salt tolerance in Arabidopsis. Plant Mol Biol. 2015;89:365–384. doi: 10.1007/s11103-015-0375-1. [DOI] [PubMed] [Google Scholar]
- 48.Hwang SG, Kim JJ, Lim SD, Park YC, Moon JC, Jang CS. Molecular dissection of Oryza sativa salt-induced RING Finger Protein 1 (OsSIRP1): possible involvement in the sensitivity response to salinity stress. Physiol Plant. 2016;158:168–179. doi: 10.1111/ppl.12459. [DOI] [PubMed] [Google Scholar]
- 49.Fang H, Meng Q, Xu J, et al. Knock-down of stress inducible OsSRFP1 encoding an E3 ubiquitin ligase with transcriptional activation activity confers abiotic stress tolerance through enhancing antioxidant protection in rice. Plant Mol Biol. 2015;87:441–458. doi: 10.1007/s11103-015-0294-1. [DOI] [PubMed] [Google Scholar]
- 50.Yan J, Wang J, Li Q, Hwang JR, Patterson C, Zhang H. AtCHIP, a U-box-containing E3 ubiquitin ligase, plays a critical role in temperature stress tolerance in Arabidopsis. Plant Physiol. 2003;132:861–869. doi: 10.1104/pp.103.020800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou J, Zhang Y, Qi J, et al. E3 ubiquitin ligase CHIP and NBR1-mediated selective autophagy protect additively against proteotoxicity in plant stress responses. PLoS Genet. 2014;10:e1004116. doi: 10.1371/journal.pgen.1004116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang W, Vinocur B, Shoseyov O, Altman A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004;9:244–252. doi: 10.1016/j.tplants.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 53.Zhu J-K. Abiotic Stress Signaling and Responses in Plants. Cell. 2016;167:313–324. doi: 10.1016/j.cell.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ohama N, Sato H, Shinozaki K, Yamaguchi-Shinozaki K. Transcriptional Regulatory Network of Plant Heat Stress Response. Trends Plant Sci. 2017;22:53–65. doi: 10.1016/j.tplants.2016.08.015. [DOI] [PubMed] [Google Scholar]