Figure 4. Inhibition mechanism of SpyCas9 by AcrIIA4.
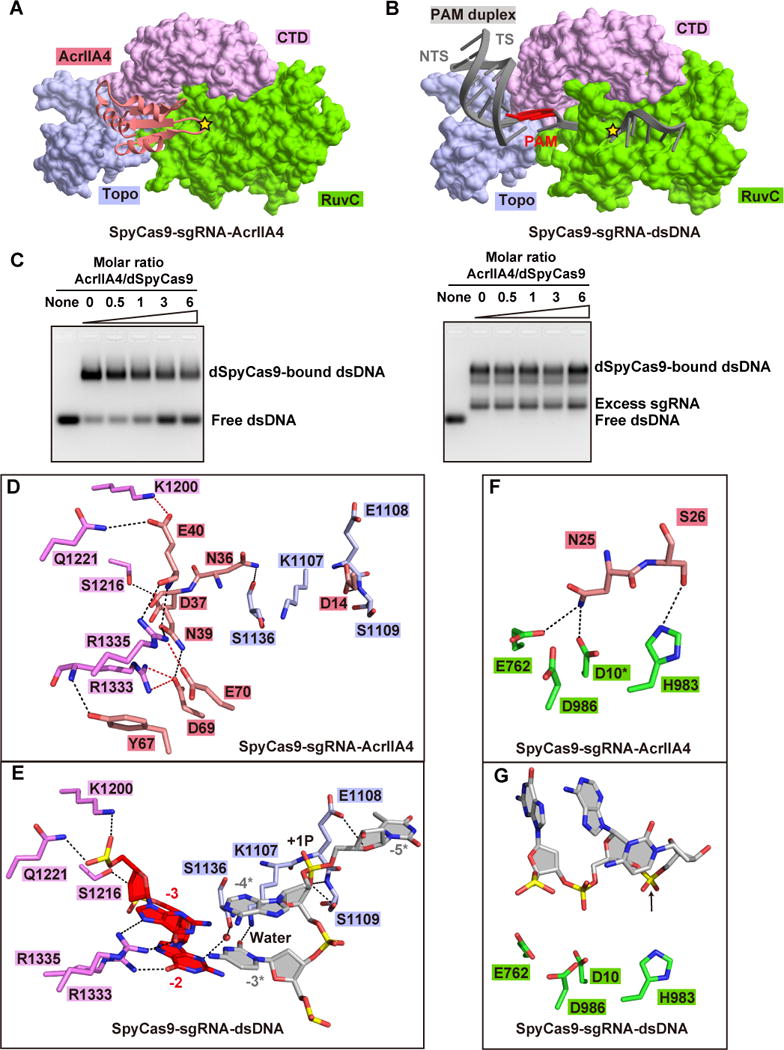
(A, B) Recognition of AcrIIA4 (panel A) and the PAM duplex (panel B) by Topo, CTD, and RuvC domains of SpyCas9. The active site of RuvC domain for non-target DNA strand in each panel is highlighted by a yellow star. TS and NTS represent target and non-target DNA strands, respectively.
(C) AcrIIA4 competitively binding to preformed SpyCas9-sgRNA binary complex (left panel) rather and SpyCas9-sgRNA-dsDNA ternary complex (right panel) in EMSA assays. The molar ratios of AcrIIA4:SpyCas9 are shown at the top of the gel.
(D, E) Detailed interactions between AcrIIA4 (panel D) and PAM duplex (panel E) with the Topo and CTD domains of SpyCas9, respectively.
(F, G) Recognition of key catalytic residues in the RuvC active site by AcrIIA4 (panel F) and non-target DNA strand (panel F). The modeled side chain of catalytic residue Asp10 is marked by asterisk. The position of cleavage site in non-target DNA strand is pointed by a black arrow.
See also Figure S4.
