Figure 5. Developmental window for effects of VIP-IN disruption.
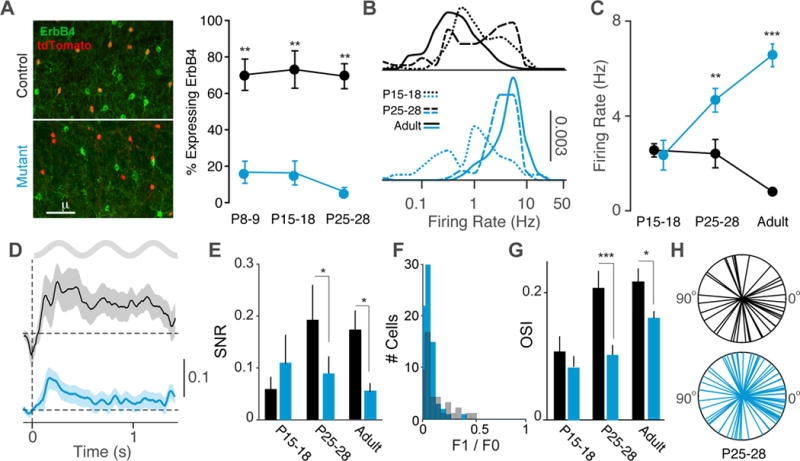
(A) ErbB4 and tdTomato immunohistochemistry in V1 cortex of control (black) and mutant (cyan) mice during postnatal development. Left: Upon Cre recombination of the reporter line Ai9, VIP cells express tdTomato. Right: ErbB4 expression in VIP interneurons quantified as a fraction of cells double-labeled for ErbB4 and tdTomato over the total number of tdTomato labeled cells, in controls (black; n = 4 mice) and mutants (cyan; n = 4 mice). (B) Distribution of firing rates of RS cells across population for three ages (P15-18, P25-28, and Adult) in controls and mutants. (C) Average firing rate of RS cells during quiescence for each age group. Controls: 113 cells, 4 mice (P15-18); 70 cells, 3 mice (P25-P28); 153 cells, 8 mice (adult). Mutants: 89 cells, 7 mice (P15-18); 103 cells, 4 mice (P25-P28); 134 cells, 8 mice (adult). (D) Average rate modulation relative to inter-trial interval around visual stimulus onset for RS cells in P25-28 animals. Controls: 43 cells, 3 mice Mutants: 74 cells, 3 mice. (E) Signal-to-noise ratio of visual responses for RS cells in controls and mutants in each age group. Controls: 23 cells, 3 mice (P15-18); 43 cells, 3 mice (P25-P28); 106 cells, 8 mice (adult). Mutants: 20 cells, 3 mice (P15-18); 74 cells, 3 mice (P25-P28); 92 cells, 7 mice (adult). (F) Histogram of F1/F0 values for RS cells in P25-28 animals. Controls: 43 cells, 3 mice. Mutants: 74 cells, 3 mice. (G) Average orientation selectivity index (OSI) of all RS cells in each age group in controls and mutants. Controls: 5 cells, 2 mice (P15-18); 42 cells, 3 mice (P25-P28); 55 cells, 7 mice (adult). Mutants: 20 cells, 3 mice (P15-18); 69 cells, 3 mice (P25-P28); 61 cells, 8 mice (adult). (H) Radial plots of preferred orientations of all RS cells in the P25-28 age group. Error bars and shading show s.e.m., p*<0.05, p**<0.01, p***<0.001.
