Figure 5. Amino Acid Sequence Alignments of Different HPV L1 Proteins and Location of Variable Regions on the Monomer Surface.
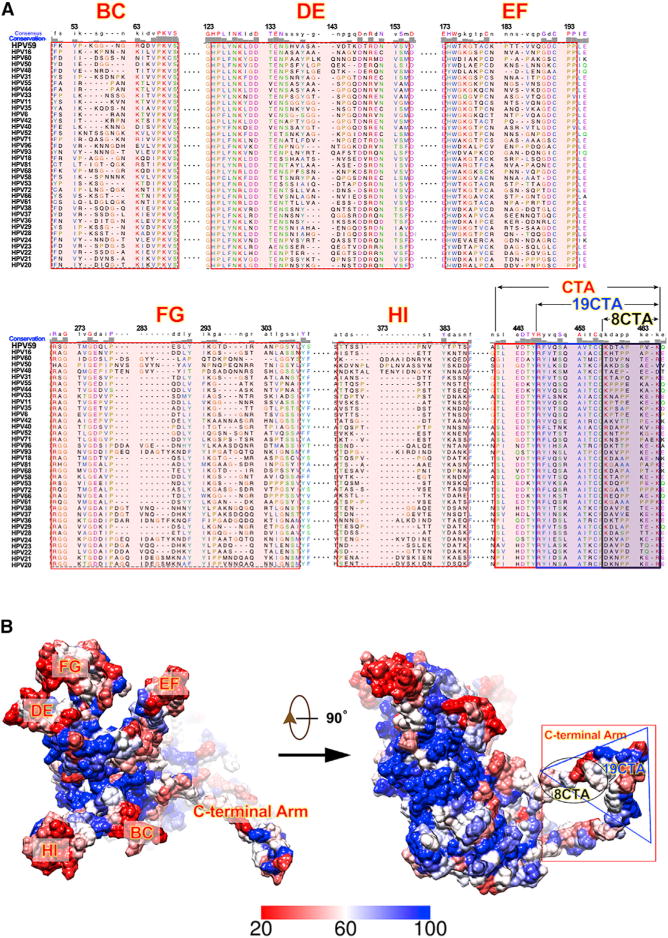
(A) Sequence alignments of the L1 proteins from 36 different HPV serotypes. The six variation regions (BC, DE, EF, FG, HI, and C-terminal arm) are highlighted in red. Alignments were determined and plotted using Chimera (Pettersen et al., 2004).
(B) Locations of regions with high sequence variation on the HPV L1 surface. The magnitude of sequence conservation based on the sequence alignments in (A) are mapped onto the surface of the HPV59 L1 monomer (left: top view; right: side view). Highly conserved regions (60%–100% conservation) are color-coded from white to blue, whereas less conserved regions (from 20% to 60%) are represented coded from red to white.
