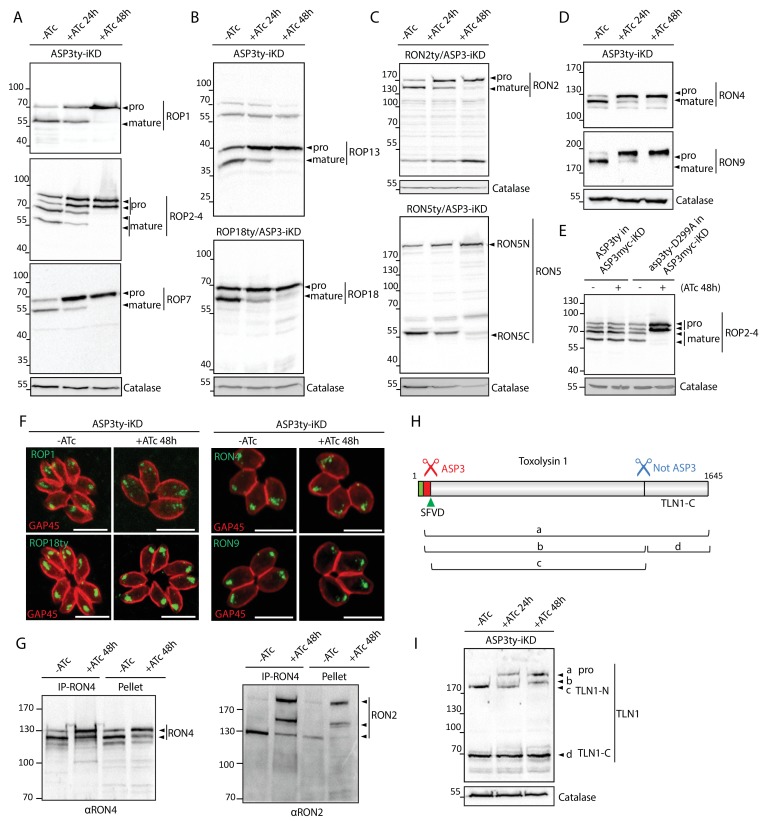
Figure 4. ASP3 is involved in the processing of rhoptry proteins.
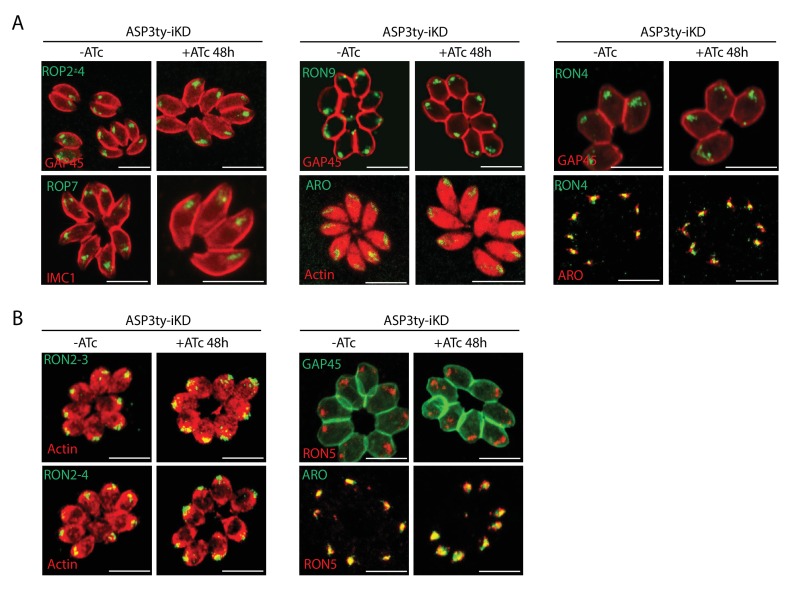
(A), (B) Immunoblots evaluating the processing of the rhoptry bulb proteins ROP1, ROP2-4, ROP7, ROP13, and ROP18 upon ASP3 knockdown. ROP18 was Ty-tagged at endogenous locus in ASP3-iKD. Catalase was used as a loading control. Arrowheads represent pro and mature forms of the proteins. (C), (D) Immunoblots evaluating the processing of the rhoptry neck proteins RON2, RON4, RON5 and RON9 upon ASP3 knockdown. RON2 and RON5 were Ty-tagged at the endogenous locus in ASP3-iKD. (E) ASP3ty but not asp3ty-D299A can rescue the processing of ROP2-4 in absence of ASP3myc. (F) IFAs evaluating localization of ROP1, ROP18, RON4, and RON9 upon ASP3 knockdown. No alteration was observed in absence of ASP3. (G) ASP3 depletion did not impact on the formation of the RON4-RON2 complex as demonstrated by co-immunoprecipitation of RON2 with RON4 antibodies. (H) Schematic for the processing events and products of TLN1. (I) Immunoblot evaluating TLN1 showed that ASP3 depletion abolished the pro-domain processing of TLN1.