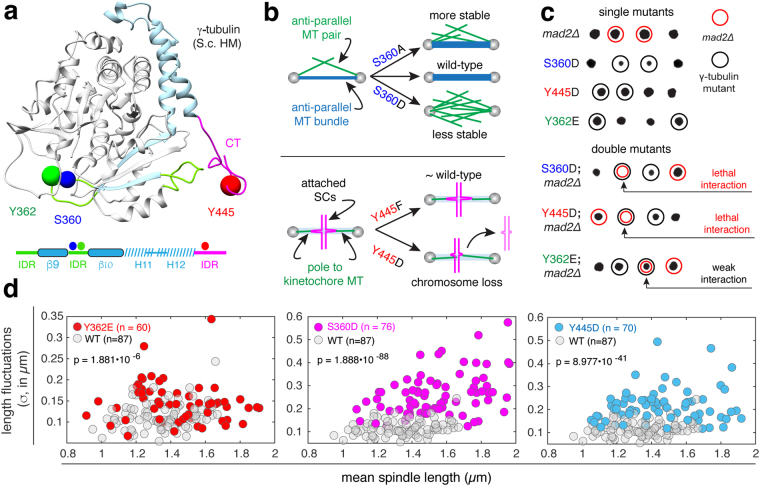
Figure 1.
Effects of γ-tubulin phosphomimetic (D/E) mutations on spindle function and spindle stability. (a) ribbon structure of yeast γ-tubulin (homology model based on the human γ-tubulin structure) with loops and structured domains in the final 132 residues shown in green and blue, the predicted intrinsically disordered (ID) c-terminus is shown in pink. Spheres (0.25 nm) indicate the positions of the side chains of residues known to be phosphorylated at SPBs in vivo; S360 (blue), Y362 (green) and Y445 (red). A schematic representing the primary sequence of the 132 residues is shown below with ID regions shown in green, the α-helices and β-strands shown in dashed blue and solid blue (respectively), and the C-terminus shown in pink. (b) Cartoon depicting the role of S360 in architecture of the anti-parallel microtubule (MT) bundle and spindle stability (left panel); schematic depicting the role of Y445 in the attachment of sister chromatids (SCs) and resulting chromosome loss (right panel). (c) Tetrad dissections of γtub-S360D, γtub-Y445D and γtub-Y362E, alone and in combination with mad2Δ. Unlike γtub-S360D and γtub-Y445D, the γtub-Y362E mad2∆ double mutant is viable, with a slight growth defect, at 25 °C. (d) Spindle length fluctuations in wild-type, γtub-Y362E, γtub-S360D and γtub-Y445D cells, plotted as a function of mean spindle length. Fluctuations for each spindle (σ) are computed as the standard deviation of length.