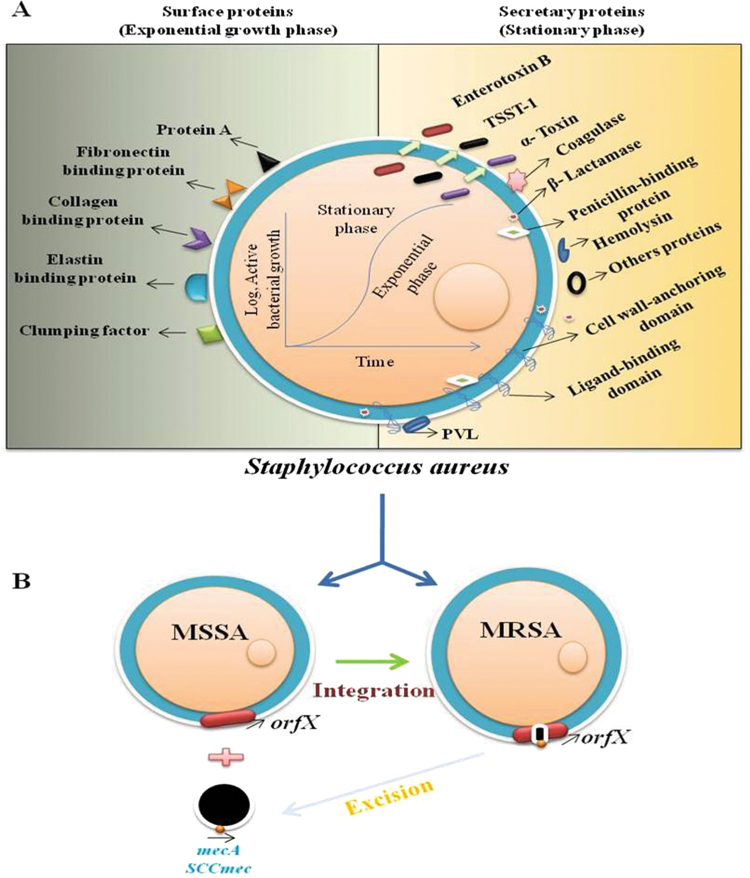
Figure 5.
Proposed study showed S. aureus associated virulence factors and resistance mechanism. (A) Number of virulence factors present on surface, secreted in exponential and stationary phase during organism life span. 1. On surface different virulence factors called adherence determinant comprises of proteins covalently anchored to cell peptidoglycans are Staphylococcoal protein A (SpA), elastin-binding protein, fibronectin-binding protein A and B (FnbpA and FnbpB), collagen-binding protein and clamping factor (Clf) A and B proteins. 2. Many group of exoproteins such as (a). secretary toxins (toxic shock syndrome toxin-1, Staphylococcal enterotoxins (SEA to SE), exfoliative toxins A and B (ETA and ETB), pyrogenic toxin superantigens (PTSAgs), α/β/γ-hemolysin, leukocidin, Panton-Valentine leukocidin (PVL) proteins vigorously stimulate T-lymphocytes to proliferate, toxic shock syndrome and food poision outbreaks) (b) enzymes including proteases, lipases, collagenase, hyaluronidase, β–lactamase, nucleases and mainly coagulase involved in converting host tissue nutrients to use for its growth, and (c) other proteins also involved in tolerating host innate and adaptive immune system including staphylococcal complement inhibitor (SCIN), staphylokinase (SAK), chemotaxis inhibitory protein of S. aureus (CHIPS), extracellular fibrinogen binding protein (Efb), extracellular adherence protein (Eap) and formyl peptide receptor-like-1 inhibitory protein (FLIPr). These multifactorial virulence factors designed in S. aureus machinery system and express at different stages of infectious conditions to act efficiently. (B) Methicillin susceptible S. aureus develop resistance to number of synthetic antibiotics by acquiring resistance machinery Staphylococcal Cassette Chromosome mec (SCCmec) harboring gene called ‘mecA’ integration to become methicillin resistant (called MRSA) and vice versa. This confirms and establish as a ‘central dogma’ to characterize strains to judge MRSA or MSSA.