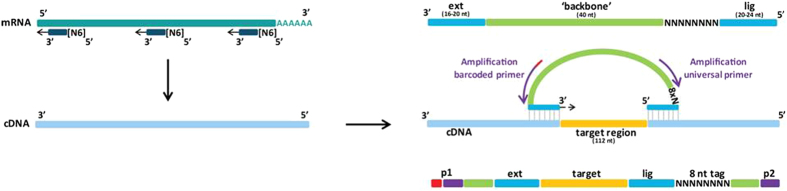
Figure 1.
Principle of smMIP-based targeted RNA sequencing. The procedure depends on the hybridization of molecular inversion probes consisting of a ligation and an extension probe that are connected via a backbone. Capture hybridization leaves for each smMIP a gap of 112 nt that is enzymatically extended and closed by ligation. After exonuclease digestion of non-ligated probes, the remaining library of circularized smMIPS is PCR-amplified with primers in the smMIP backbone. Note that the ligation probe is flanked by a random 8N sequence that allows correction for PCR duplicates. During PCR, for each sample a unique barcode primer is used allowing identification of sample-specific reads.