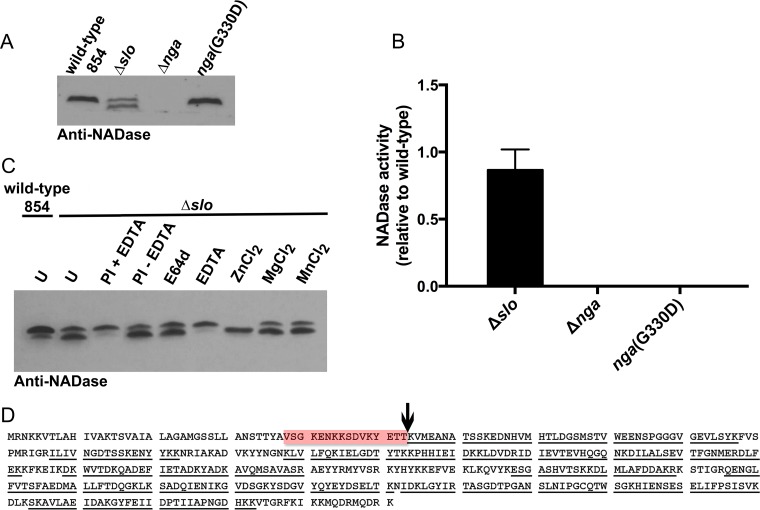
FIG 3 .
SLO prevents proteolytic cleavage of NADase. (A) Western blot using antiserum to NADase revealed two distinct bands in culture supernatant of a mutant strain deficient in SLO production (Δslo) but not in supernatants from the wild type or a mutant producing NADase G330D. (B) NADase enzymatic activity associated with the Δslo mutant was not significantly different from that of wild-type 854 as determined by one-way analysis of variance with Tukey’s posttest. (C) NADase cleavage in the Δslo mutant is zinc dependent; it is inhibited by EDTA and enhanced by the addition of ZnCl2 (1 mM) but not MgCl2 (1 mM) or MnCl2 (1 mM). Experiments were performed three times, and where appropriate, the mean ± standard deviation is represented (U, untreated; PI, protease inhibitor). (D) The NADase cleavage site in the Δslo mutant was determined by extraction of full-length and processed NADase from SDS-PAGE and analysis by tryptic digest and LC–MS-MS for fragment identification. The processing site was found to be between amino acids T53 and K54 (vertical arrow). The region of the protein identified in the full-length NADase but not the processed form is highlighted in red. Underlining denotes peptides identified in the analysis for both forms of NADase.