Fig. 1.
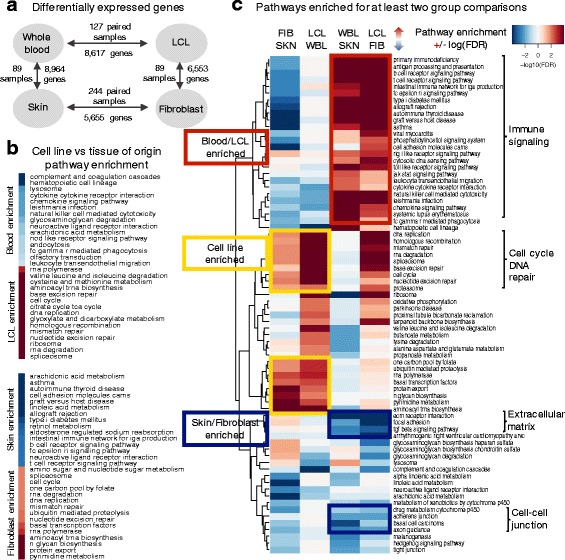
Pathways are differentially expressed between cell lines and their tissues of origin. a Number of differentially expressed genes (absolute log2 fold change >2 and FDR < 0.05) using voom on paired samples. b Results of GSEA reported based on the log10(FDR) significance scale, with one group in red and the other one in blue. The 15 pathways most significantly differentially expressed between each cell line and its tissue of origin. c Pathways enriched for at least two group comparisons (FDR < 0.05). The pathways differentially expressed between the tissues that are also differentially expressed between the cell lines (preserved pathways) are highlighted in red and blue. Pathways over-expressed in both cell lines compared to their tissues of origin are highlighted in yellow. Rows are ordered by hierarchical clustering of the enrichment significance values, log10(FDR). To represent the FDR significance in the heatmap, the color was saturated at 1.1 × 10−4. The exact reported FDR can be found in Additional file 2
