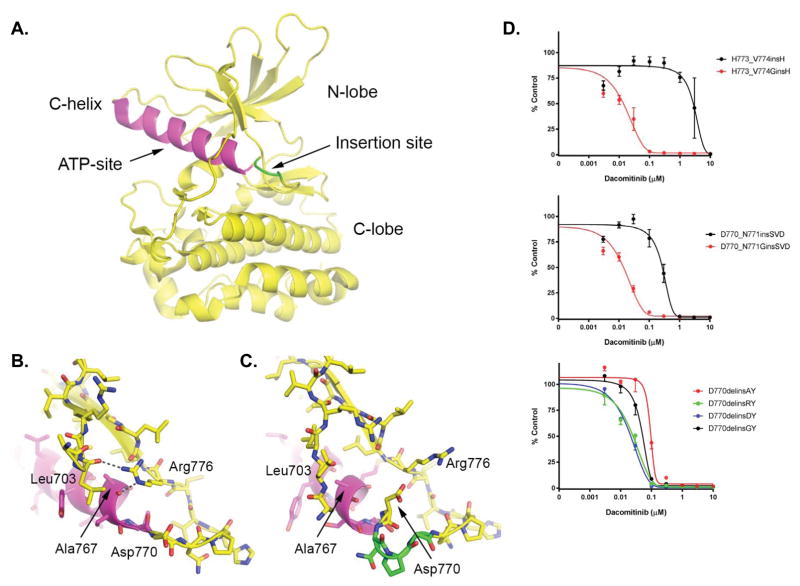
Figure 2.
Structural context of exon20 insertion mutants and impact on dacomitinib sensitivity. A. Previously determined structure of the EGFR insNPG exon20 insertion mutant (PDB ID: 4LRM), highlighting the architecture of the kinase domains and the typical location of exon20 insertion mutants. The insNPG insertion is highlighted in green and the C-helix is shaded magenta. The ATP-site, which is also the site of binding of inhibitors, lies between the N- and C-lobes of the kinase as indicated. B. In the inactive conformation of the EGFR kinase, Arg 776 hydrogen bonds with the carbonyl of Ala767 at the end of the C-helix, and with Leu703 in the N-terminal portion of the kinase. We propose that this interaction is important for the stability of the inactive conformation of the kinase, as well for the transition between the inward (active) and outward (inactive) positions of the C-helix which may in turn control access of inhibitors to the adjacent ATP-site. (Panel drawn from PDB ID 1XKK.) C. Detailed view of the insertion site in the insNPG mutant (PDB ID 4LRM), colored as in panel A. The insertion reorients Asp770 such that it blocks access of Arg776 to the end of the C-helix. Other inhibitor-resistant mutants may have a similar effect on the position of Asp770, but at present crystal structures are available only for insNPG. In all of the inhibitor-sensitive insertion mutants studied here, a glycine residue is inserted in place of Asp770, potentially restoring the ability of Arg776 to rearrange in communication with the C-helix. D. Ba/F3 cells expressing different modified exon 20 insertion mutations. In the top two, Asp770 has ben replaced by a Gly. In the bottom, Gly770 has been replaced by either Asp, Ala or Arg. Cells were treated with dacomitinib at the indicated concentrations, and viable cells were measured after 72 hours of treatment and plotted relative to untreated controls.